b. University of Chinese Academy of Sciences, Beijing, 100049, China
Transitioning at the right time from vegetative to reproductive growth, i.e. flowering time, is essential for maximizing fitness. It has been well established that flowering transition is tightly regulated by an intrinsic and complex gene-regulatory-network (GRN) by incorporating endogenous and exogenous signals including nutrients like nitrogen (Andres and Coupland, 2012, Marin et al., 2011, Michaels, 2009, Nee et al., 2017, Stitt, 1999). Nitrate (NO3-), a major nutrient taken up mainly via lateral roots, has been considered as one of the signaling molecules for plant development (Bouguyon et al., 2012, Stitt, 1999). Nitrogen deficiency often induces early flowering, a pattern identified more than a hundred years ago (Klebs, 1913). Furthermore, high levels of nitrogen normally delay flowering time, likely due to the high rates of photosynthetic CO2 fixation, which balance nitrogen assimilation (Bernier et al., 1993, Noguero and Lacombe, 2016). A U-shaped flowering curve has been observed in Arabidopsis accession Col-0, in which both low and high concentrations of nitrate delay flowering while only plants growing under optimal nitrate supply conditions flower early (Lin and Tsay, 2017, Marin et al., 2011, Yuan et al., 2016).
The flowering time GRN involves several genetic pathways, namely autonomous and vernalization, aging (including sugar-mediated), gibberellin acid, and circadian clock and photoperiod pathways, each featuring several key transcription factors (Andres and Coupland, 2012, Michaels, 2009, Nee et al., 2017, Srikanth and Schmid, 2011). SOC1 and FT are among the key transcriptional regulators that integrating signals from almost all the genetic pathways and from exogenous environmental stress signals like drought or nutrient status variation (Conti, 2019, Hwang et al., 2019, Kant et al., 2011, Liu et al., 2013, Olas et al., 2019, Riboni et al., 2013, Riboni et al., 2016). Almost all these genetic pathways have been implicated in the regulation of nitrate-mediated flowering (see reviews Fredes et al., 2019, Lin and Tsay, 2017, Weber and Burow, 2018). GI has been considered as one of the key factors downstream of circadian clock and photoperiod pathway to tune the expression of FT directly or indirectly via CO. Depleting simultaneously the photoperiod and gibberellin acid pathways as well as autonomous regulators causes a never-flowering phenotype, which can be recovered by a low-nitrate induction or vernalization treatment (Marin et al., 2011, Reeves and Coupland, 2001). Recently, the AP2-domain transcription factor NGR5 is revealed to recruit polycomb repressive complex 2 in a nitrogen-dependent manner to modulate the H3K27me3 modification level of chromatin, and compete with DELLA proteins, key repressors in gibberellin acid signaling pathway, for regulation of the activity of downstream genes (Wu et al., 2020).
Nitrate supply significantly affects the expression of aging pathway genes, for example the miR156-SPLs and their downstream miR172-AP2s modules (Fischer et al., 2013, Gras et al., 2018, Liang et al., 2012, Olas et al., 2019, Pant et al., 2009, Srikanth and Schmid, 2011, Wang et al., 2009, Wu and Poethig, 2006). Nitrate appears to act upstream of aging pathway, as SPL3, SPL4 and SPL5 promoters contain many nitrate-responsive elements (NREs) that can be bound by NLP6 and NLP7 (Konishi and Yanagisawa, 2013, Olas et al., 2019). However, low-nitrate-mediated flowering delay under short-day conditions relies on the accumulation of the carbon signaling molecule, trehalose-6-phosphate (T6P), indicating that the interactions between nitrate and sugar signaling jointly play an important role in setting flowering time of Arabidopsis (Olas et al., 2019). Interestingly, T6P signaling also works on the miR156-regulated SPLs to tune flowering time (Wahl et al., 2013).
Previous studies have shown how nitrogen, both as an essential macronutrient and a signaling molecule, regulates flowering time in Arabidopsis and the monocot model rice (Wang et al., 2018). However, only limited information can be found for nitrate-mediated flowering time variation in naturally occurring accessions of Arabidopsis (de Jong et al., 2019, Marin et al., 2011). In this study, we used three Arabidopsis accessions to investigate the effect of nitrate supply change on flowering time variation in long-day and day-neutral conditions. We reveal a accession-dependent flowering time variation upon changing of nitrate availability, and this associates with the accession-specific expression of key flowering-time genes. These findings provide new insights on nitrate-mediated flowering time regulation.
2. Materials and methods 2.1. Plant materials and growth conditionsArabidopsis thaliana accessions of Col-0, Ler (Landsberg erecta) and Ws (Wassilewskija) were kindly provided by Prof. Maarten Koornneef. The single-seed propagated and harvested seeds for all three accessions in the same batch were used for flowering time assays. Arabidopsis seeds were sterilized, stratified, and sown on agar plates according to procedures described in Hu et al. (2014). Agar plates were prepared according to recipes described by Lin et al. (2017) (Table S1). On each plate (130 × 130 mm square plate), four seeds of each accession (in total 12 seeds) were sown with positions randomly distributed. Plants were grown in chambers (Percival) under long-day (LD: 16-h light and 8-h night at 21 ℃) and day-neutral (MD: 12-h light and 12-h night at 21 ℃) conditions. To minimize positional effects during growth, plates growing vertically were randomized every two days.
2.2. Flowering time scoring and measurement of other traitsFlowering time assays were performed on above-mentioned agar plates supplied with different levels of KNO3 (0.1, 0.5, 1, 2, 4, 8, 16, 32 mM). A pilot experiment scoring both rosette leaf number and the days after germination (DAG) upon flowering suggested DAG was the more reliable way to monitor flowering time (data not shown); therefore, we traced in each trial for each line at each nitrate concentration at least 15 plants growing on at least four plates for flowering time.
We also measured the primary root length, total number of lateral roots longer than 3 mm (that assumed to function normally in nutrient assimilation), and fresh weight of aerial part for plants growing on different concentrations of KNO3 with minimum 21 plants examined. Seedlings at DAG14 in both LD and MD were used for these measurements.
Statistical analysis of trait variation was carried out with Student's t-test in excel and Pearson's correlation test in R.
2.3. RNA extraction and gene expression assaysTotal RNA extraction and reverse transcription followed by quantitative PCRs were carried out according to Hu et al. (2014). Seedlings at DAG15 grown under both LD and MD conditions were harvested 30 min prior to dawn and used for RNA extraction. qRT-PCR was conducted with gene-specific primers (but could amplify both Col-0 and Ws accessions) using PP2A as references. See Table S2 for detailed information on primers used in this study. For each trial and each line at each nitrate concentration, three biological replicates (each contained at least 6 seedlings from three plates) were used for quantification with each having three technical replicates. Gene expression assays were duplicated and both showed the same pattern, with only data from one trial shown in Fig. 3, Fig. 4, Fig. 5. Statistical differences were compared with Student's t-test.
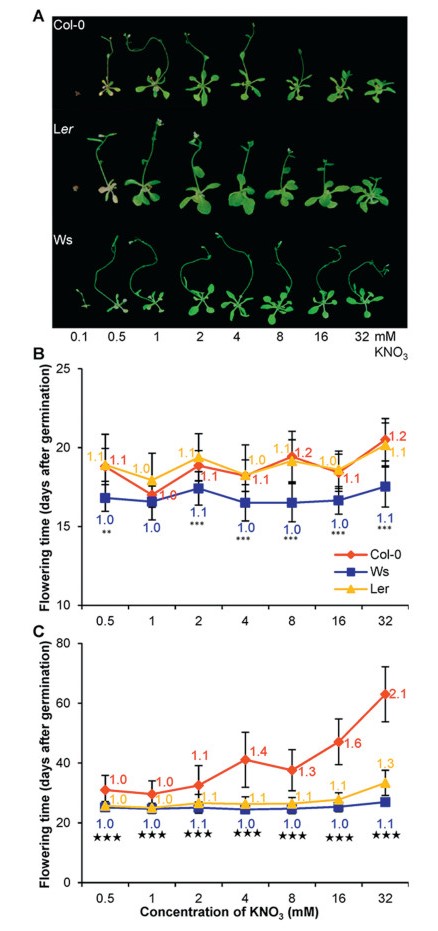
|
| Fig. 1 Natural variation in flowering time responds to nitrate fluctuation (KNO3 in mM) under both long-day (LD; 16 h/8 h day/night) and day-neutral (MD; 12 h/12 h day/night) conditions. A. Photograph of representative plants growing under LD conditions for 20 days after germination (DAG). The upper, middle, and lower panels show the morphology for Col-0, Ler, and Ws plants, respectively. Note that plants grown at 0.1 mM concentration of KNO3 display a strong stress phenotype. B. Flowering time in DAG for plants growing under LD conditions. For all three accessions, numbers in corresponding colors indicate the fold change taking the flowering time at 1 mM as 1 (the same for C). C. Flowering time in DAG for plants growing under MD conditions. For B and C, * and ★ indicate the significances between Col-0 and Ws at each nitrate concentration under LD (*) and MD (★) conditions, respectively. ***/★★★, P < 0.001, and **/★★, P < 0.01. See Fig. S 1 for other comparisons. |
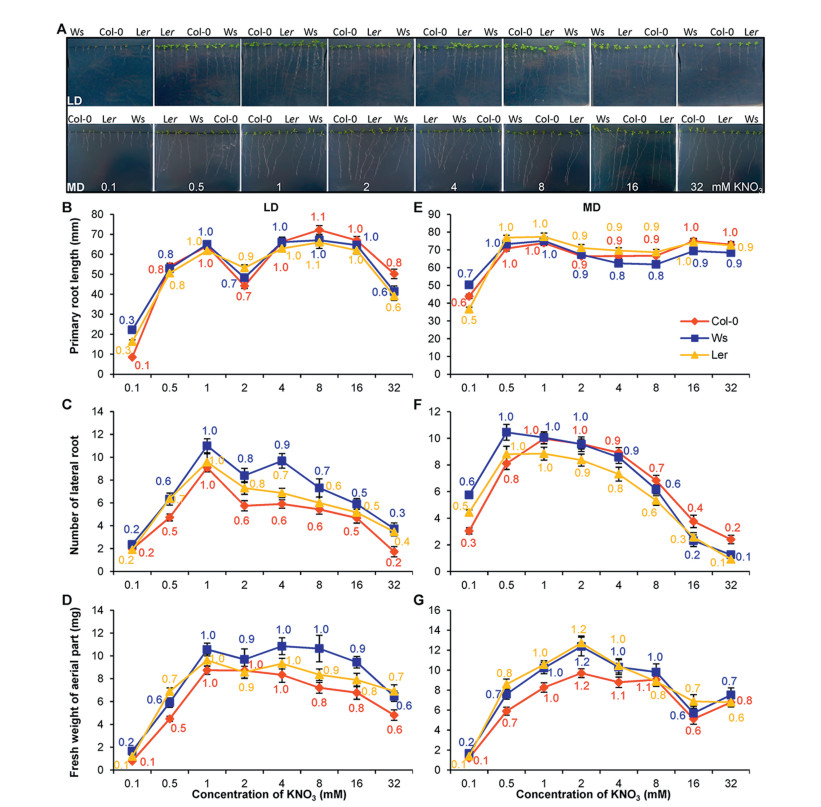
|
| Fig. 2 Effects of nitrate fluctuation on flowering time variation anti-associates partially with growth phenotypes. A. Photograph of representative plants growing under LD (upper panel) or MD (lower panel) conditions for 14 DAG. For each plate, seeds of the three accessions (Ws, Col-0, and Ler) were randomly sown to minimize positional effect. Note the growth of plants at 0.1 mM concentration shows a strong stress phenotype. B-G. The primary root length (B and E), number of lateral roots (C and F), and fresh weight of aerial part of seedlings (D and G) for the three accessions grown under different nitrate concentrations (X-axis) at LD (B, C, and D) and MD (E, F, and G) conditions. Bars show the standard deviation of means for at least 15 plants of each accession. For C and F, only lateral roots longer than 3 mm are counted. For B to G, numbers in corresponding colors indicate the fold change taking the trait mean value at 1 mM as 1. This experiment was duplicated and both gave similar results. |
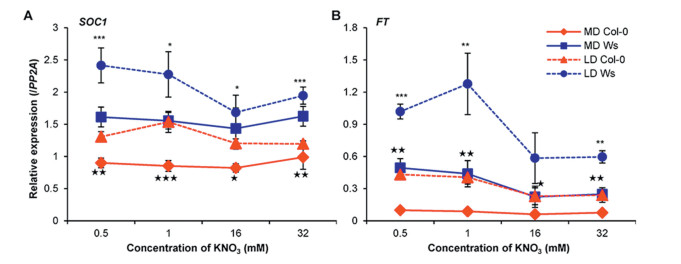
|
| Fig. 3 Natural variation in relative expression of SOC1 (A) and FT (B) in response to nitrate concentration fluctuation (X-axis) is associated with flowering behaviors of Col-0 (red) and Ws (blue) under LD (broken lines) and MD (solid lines) conditions. Quantitative RT-PCR was used to quantify the relative expression in three biological replicates for 15 DAG seedlings with standard deviation of means shown. Note that, in comparison to Col-0, Ws features a relatively higher basal expression for both genes and for both growth conditions. In both A and B, * marks significance levels under LD condition between genotypes: ***, P < 0.001, **, P < 0.01, and *, P < 0.05; ★ shows significant difference under MD condition, ★★★, P < 0.001, ★★, P < 0.01, and ★, P < 0.05). |
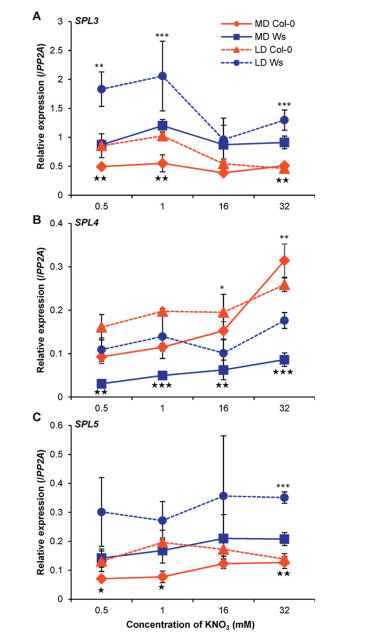
|
| Fig. 4 Relative expression of aging pathway genes SPL3 (A), SPL4 (B), and SPL5 (C) in Col-0 (red) and Ws (blue) seedlings grown on different nitrate concentrations under LD (broken lines) and MD (solid lines) conditions. Mean values with standard deviation of three biological replicates are shown. * marks significance levels under LD condition between genotypes: ***, P < 0.001, **, P < 0.01, and *, P < 0.05; ★ shows significant difference under MD condition, ★★★, P < 0.001, ★★, P < 0.01, and ★, P < 0.05. |
|
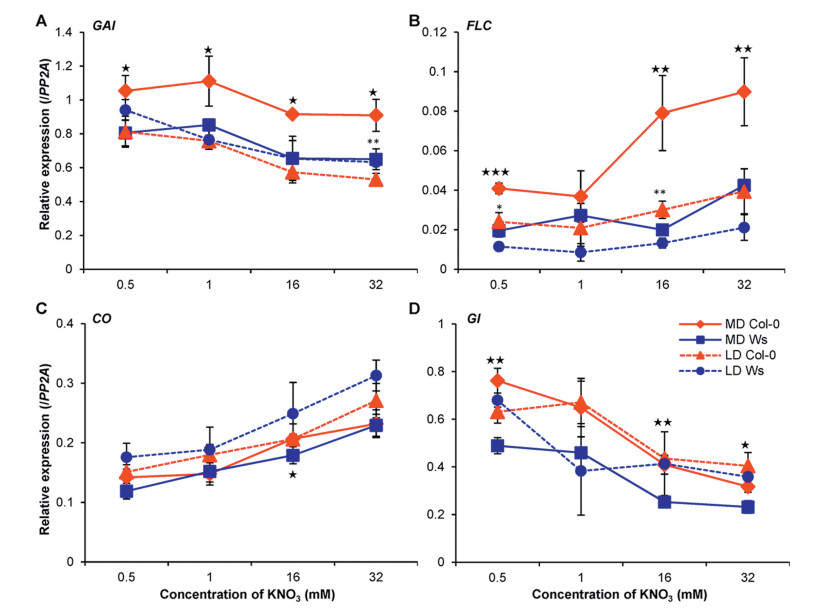
| Fig. 5 Relative expression of GAI (A), FLC (B), CO (C) and GI (D) in Col-0 (red) and Ws (blue) seedlings grown on different nitrate concentrations under LD (broken lines) and MD (solid lines) conditions. Mean values with standard deviation of three biological replicates are shown. * marks significance levels under LD condition between genotypes: ***, P < 0.001, **, P < 0.01, and *, P < 0.05; ★ shows significant difference under MD condition, ★★★, P < 0.001, ★★, P < 0.01, and ★, P < 0.05. |
To explore whether flowering time of different genotypes can respond to nitrate availability, three frequently used accessions of Arabidopsis thaliana, Col-0, Ler, and Ws, were used to test their flowering time behaviors upon growing on agar plates (see Table S1 for chemical components) supplied with various concentrations of KNO3 (0.1, 0.5, 1, 2, 4, 8, 16, and 32 mM). We used days after germination (DAG) upon flowering to monitor flowering time because rosette leaf production could be significantly modified by nitrate supply (Lin and Tsay, 2017, Marin et al., 2011, Olas et al., 2019, Tschoep et al., 2009), a similar pattern also observed in our pilot experiments (data not shown).
Plants of all three accessions growing in 0.1 mM KNO3 medium displayed a strong stress symptom (e.g., chlorotic leaves or yellowish/brownish small seedlings; Fig. 1A and Fig. S1), though Ws plants were able finally to bolt under long-day (LD; 16 h/8 h, light/night) conditions (both Col-0 and Ler did not flower). To avoid masking by N-dependent responses (Tschoep et al., 2009), we did not score flowering phenotypes for any accession grown in 0.1 mM KNO3. Under LD conditions, Col-0 showed a fluctuation with its flowering time from 17 DAG (1 mM) to 18.8 DAG (0.5 mM; about 1.1-fold in comparison to 1 mM concentration) or 20.5 (32 mM; 1.2-fold) DAG (Fig. 1B and Fig. S1). Nitrate concentration at 1 mM seemed being the optimal conditions for Col-0 to flower with both lower and higher nitrate delaying floral transitions. Ler also showed a dynamic flowering behavior, though to less extent of variation comparing to Col-0, from 17.9 DAG at 1 mM to 18.9 DAG (0.5 mM) or 20.2 (32 mM; ~1.1 fold) DAG. However, Ws featured a much earlier but less variable flowering on average (16.6-17.5 DAG). This indicates that these accessions do respond differentially in their flowering time to fluctuation of nitrate availability with Ws the less and Col-0 the most variable, corroborating with a previous notion that Arabidopsis accessions have different plasticity of flowering in responding to nitrate concentrations (de Jong et al., 2019).
3.2. Photoperiod affects accession-specific flowering time variation in responding to nitrate supplyWe next tested whether photoperiod had any influence on flowering time variation upon nitrate fluctuation for different genotypes, as it has been established that photoperiod can modulate the flowering time of Col-0 upon nitrate supply changes (Olas et al., 2019, Weber and Burow, 2018). All plants of each accession flowered significantly later when grown under day neutral (MD) conditions than LD conditions (Fig. 1C and Fig. S1). Both Ws and Ler displayed very little variation (~1.0- to 1.3- fold in comparison to the flowering time at 1 mM) in their flowering time when grown on different concentrations of KNO3. In contrast, Col-0 plants varied considerably, from 31 DAG at 0.5 mM (~1.0-fold in comparison to the flowering time at 1 mM) to 63 DAG at 32 mM (~2.1-fold). These data clearly suggest a significant influence of photoperiod on accession-specific flowering time variation upon nitrate change in Arabidopsis. Col-0 is more sensitive to nitrate supply levels than Ler and Ws, consistent with a previous finding that most flowering time variation caused by nitrate supply change is due to genetic differences among lines (de Jong et al., 2019, Pigliucci and Schlichting, 1995).
| Accession | Parameter | LD condition | MD condition | |||||
| Primary root length | Number of lateral root | Fresh weight of aerial part | Primary root length | Number of lateral root | Fresh weight of aerial part | |||
| Col-0 | cor | -0.36 | -0.91 | -0.64 | 0.32 | -0.9 | -0.38 | |
| t | -0.85 | -5.03 | -1.84 | 0.76 | -4.54 | -0.901 | ||
| df | 5 | 5 | 5 | 5 | 5 | 5 | ||
| p | 0.43 | 0.004 | 0.13 | 0.48 | 0.006 | 0.407 | ||
| Ws | cor | -0.96 | -0.53 | -0.55 | 0.18 | -0.72 | -0.49 | |
| t | -7.4 | -1.38 | -1.48 | 0.407 | -2.32 | -1.24 | ||
| df | 5 | 5 | 5 | 5 | 5 | 5 | ||
| p | 0.001 | 0.226 | 0.199 | 0.701 | 0.068 | 0.269 | ||
| Ler | cor | -0.76 | -0.76 | -0.71 | -0.17 | -0.85 | -0.58 | |
| t | -2.61 | -2.579 | -2.2672 | -0.39 | -3.64 | -1.63 | ||
| df | 5 | 5 | 5 | 5 | 5 | 5 | ||
| p | 0.048 | 0.049 | 0.073 | 0.71 | 0.15 | 0.164 | ||
We continued to investigate whether the flowering variation in these accessions correlates with growth patterns. We measured the primary root length, total number of lateral roots longer than 3 mm, and fresh weight for plants growing under both LD and MD for 15 DAG (Fig. 2). All three accessions showed a relatively stable and identical pattern of primary root length for plants grown under both LD and MD conditions, except at 0.1 mM KNO3, where an obvious reduction was observed (Fig. 2A, B and E). Lateral root number and fresh weight of aerial parts for all three accessions displayed an "inverted U type", with strong variation especially at concentrations of 0.1 and 32 mM under both photoperiod rhythms. LD growing Ws plants had more lateral roots and fresh weight of aerial parts than Col-0 and Ler (Fig. 2C, D, F and G). These data indicate that nitrate supply affects growth traits in an accession-dependent manner with the best growth occurring around 1 mM KNO3. Furthermore, photoperiod seems to affect only the absolute levels of primary root length, but does not impact the number of lateral roots or fresh weight of aerial part of these accessions.
Interestingly, only Col-0 plants showed an anti-correlation pattern between flowering time and lateral root number when grown in both LD (Pearson's correlation test, cor = -0.91, p = 0.004) and MD (Pearson's correlation test, cor = -0.9, p = 0.006) conditions (Table 1). Ler displayed a negative association between flowering time and number of lateral roots only in LD conditions (cor = -0.76, p = 0.049). We observed a negative correlation between flowering time and primary root length for both Ws (cor = -0.96, p = 0.001) and Ler (cor = -0.76, p = 0.048) plants grown under LD but not MD conditions. These data clearly indicate that accession-specific flowering time variation responds to nitrate supply change only under certain photoperiod conditions.
We next examined whether these genotype-specific flowering time behaviors are associated with variation in gene expression in key flowering integrators.
3.4. Genotype-specific expression of SOC1 and FT underlies flowering time variationSOC1, but not FT, has been considered as one of the key factors regulating flowering time in shoot apical meristem under low or optimal nitrate conditions (Olas et al., 2019). Interestingly, corroborating with the earlier flowering phenotype under both LD and MD conditions, SOC1 displayed a constant higher expression at all four nitrate concentrations in Ws plants than Col-0 plants (Fig. 3A). Nitrate only mildly modulated the SOC1 expression levels in both lines under both photoperiod conditions.
FT, transcribed in leaf phloem, is a mobile signal moved to shoot apical meristem to activate downstream flowering promoters like SOC1, AP1 and several other transcription factors (Bratzel and Turck, 2015, Turck et al., 2008). Because photoperiod significantly modulated flowering time in our experiments (Fig. 1), we next examined FT expression levels in Col-0 and Ws plants following variation in both nitrate supply and photoperiod conditions (Fig. 3B). In general, FT expression showed a highly similar pattern to that of SOC1 for both genetic materials under both photoperiod conditions, indicating that both FT in leaf and SOC1 in shoot apical meristem are underlying the line-specific flowering time variation responding to nitrate variation. Interestingly, we did not detect any nitrate-regulatory-element (NRE) change in the promoter regions for both FT and SOC1 of both Col-0 and Ws (data not shown), therefore, we next looked for potential upstream regulators associating with expression variation.
3.5. Diversified relationships between expression of SPLs and flowering time variationWe first examined the miR156-SPLs expression in the aging pathway of flowering time regulation (Srikanth and Schmid, 2011, Wang et al., 2009, Wu and Poethig, 2006). miR156-SPLs expression can be modified significantly by low nitrate supply, thus has the potential to influence floral transition timing under altered nutrient conditions (Fischer et al., 2013, Liang et al., 2012, Olas et al., 2019, Pant et al., 2009). When grown under LD conditions, both Col-0 and Ws featured a relatively repressed expression of SPL3 with higher nitrate supplies (16 and 32 mM in contrast to 0.5 and 1 mM) (Fig. 4A). SPL5 expression did not change in response to different nitrate levels in either accession under LD or MD conditions. However, SPL3 and SPL5 expression was relatively higher in Ws plants than in Col-0 plants (Fig. 4C), which is consistent with their flowering time variation (Fig. 1). In our analysis under both LD and MD conditions, Ws exhibited a relatively higher expression of both SPL3 and SPL5 but a relatively lower expression of SPL4 (Fig. 4B). Furthermore, SPL4 expression of Col-0 increased about 2.5-fold at 32 mM (compared to 1 mM) KNO3 concentration, which is contradicting to the flowering time dynamics between these two accessions (Fig. 1). Consistent with previous report using Col-0 (Olas et al., 2019), SPL4 expression did not change in response to fluctuation of nitrate concentrations under LD conditions. Our data therefore indicate that these three SPLs may differ between Ws and Col-0 in specifying the flowering time variation upon fluctuation of nitrate supply.
3.6. Expression of FLC associates with genotype-specific flowering time variation upon alteration of nitrate supplyingEndogenous GA levels can be significantly modulated upon growing on low or high nitrate medium, and nitrate levels can also regulate the expression of genes in GA-mediated flowering pathway (Liu et al., 2013), though the loss-of-function mutants gai or ga1-3 in Ler background only showed a mild change in nitrate-mediated flowering time variation (Marin et al., 2011). We found that, in Col-0 and Ws, GAI expression decreased slightly when nitrate concentration increased under MD conditions (Fig. 5A), indicating that nitrate fluctuation has little impact on variation of GAI expression. However, we cannot exclude the possibility that other components in the GA-mediated flowering time network may participate in the regulation of the accession-specific flowering time variation upon nitrate change.
However, FLC seems to underlie the flowering time variation in that its expression increased for about 2.1 (16 mM) to 2.4 (32 mM) fold in Col-0 under MD conditions, while its expression did not change for Ws (both LD and MD conditions) or Col-0 under LD conditions (Fig. 5B). Not surprisingly, nitrate-mediated flowering time variation correlates well with FLC expression change in Col-0 (Kant et al., 2011). Because all three accessions do not feature a functional FRI, a strong promoter of FLC expression (Johanson et al., 2000, Korves et al., 2007, Stinchcombe et al., 2004), it will be therefore very interesting to compare the flowering time variation upon nitrate supply change in natural accessions with functional FRI.
3.7. Dynamic expression of GI, not CO, is linked to FT expressionOur results indicate that photoperiod may play an essential role in the variation of nitrate-mediated genotype-specific flowering time. Nitrate availability can significantly alters the expression of major photoperiod pathway genes like CO, CRY1 and genes in circadian clock oscillators, and while loss-of-function or overexpression of part of these genes leads to obvious flowering time change (Marin et al., 2011, Weber and Burow, 2018, Yuan et al., 2016). Furthermore, photoperiod pathway seems to interact with the gibberellin acid and autonomous pathways to modulate nitrate-regulated floral transition (Marin et al., 2011). Indeed, the expression of CO in both Col-0 and Ws plants responded to increase of nitrate concentration in the medium, though no difference could be observed between accessions (Fig. 5C), indicating that CO expression was unlikely associated with line-specific flowering time regulation. However, we found the expression of another important player, GI, one key player in photoperiod and circadian clock pathway, displayed a correlation with floral transition difference between Col-0 and Ws, especially under MD conditions (Fig. 5D). GI can regulate the FT expression either directly by binding to the FT promoter, or indirectly by modifying CO transcription, by modulating the activity of miR172/AP2-like modules, or by CDFs-mediated transcriptional regulation (Mishra and Panigrahi, 2015). However, GI can also modulate CO protein stability via physical interaction with FKF1 and ZTL, and thus indirectly repress FT expression (Song et al., 2014). Interestingly, we detected a relatively lower expression of GI in Ws plants than Col-0 under MD conditions (Fig. 5D). These data suggest that the accession-specific expression of FT can be modified by GI, independent of CO transcription, or other players in the photoperiod and circadian clock pathway, to time the floral transition upon nitrate fluctuation.
4. ConclusionIn this study, we show that the flowering time variation in responding to nitrate supply fluctuation is genotype-specific in A. thaliana, a pattern also observed previously (de Jong et al., 2019, Marin et al., 2011). Differences in flowering time responses seem depending on the light rhythm that the accessions are growing. However, accession-specific effects of flowering time variation anti-correlates with some growth traits, for example lateral roots number. We demonstrate that the flowering time variation in Arabidopsis accessions can attribute to the expression dynamics of integrators (e.g., FT and SOC1) upon change of nitrate supplies. Although the potential molecular mechanisms still await further investigation, we illustrate that expression change of specific transcription factors in the photoperiod (GI), autonomous (FLC), and aging (SPLs) pathways and/or their interactions correlate well with expression variation of floral integrators, thus might contribute to the genotype- and photoperiod-specific flowering time responses to nitrate supply change. Nitrate serves as both nutrients and signaling molecules regulating many aspects of plant developmental and environmental adaptation processes. Therefore, our efforts shed light on the dissection of molecular genetic basis on how flowering time and nitrate assimilation are coordinately regulated during plant life-cycle. Furthermore, this study provide novel insights into the breeding for crops requiring optimal but relatively lower nitrate supply in agriculture, one of the most desirable goals in agricultural efforts (Li et al., 2017).
Author contributionsJ-Y H designed the research; F-H Y, L-P Z, F C, and D-M Y performed the research; J-Y H and F-H Y analyzed data with the help from other authors; J-Y H wrote the paper with the help from F-H Y and other authors. All authors read and approved the contents in this manuscript.
Declaration of Competing InterestThe authors declare no conflict of interest.
AcknowledgementsWe thank Yibo Sun, Dan Wang, Shulan Chen for their assistance in experiments. We appreciate Prof. Shibao Zhang for useful discussion. This work was supported by grants from National Natural Science Foundation of China (31570311 to J-Y H and 31800261 to F C), from the CAS Pioneer Hundred Talents Program (292015312D11035 to J-Y H), and CAS Key Laboratory for Plant Diversity and Biogeography of East Asia to J-Y H, from the Postdoctoral targeted funding from Yunnan Province and the Yunnan basic and applied research funding to F C. This work is partially facilitated by the Germplasm Bank of Wild Species of China. The authors declare no conflict of interest.
Appendix A. Supplementary dataSupplementary data to this article can be found online at https://doi.org/10.1016/j.pld.2020.05.004.
Andres F., Coupland G., 2012. The genetic basis of flowering responses to seasonal cues. Nat. Rev. Genet, 13: 627-639. DOI:10.1038/nrg3291 |
Bernier G., Havelange A., Houssa C., et al, 1993. Physiological signals that induce flowering. Plant Cell, 5: 1147-1155. DOI:10.2307/3869768 |
Bouguyon E., Gojon A., Nacry P., 2012. Nitrate sensing and signaling in plants. Semin. Cell Dev. Biol, 23: 648-654. DOI:10.1016/j.semcdb.2012.01.004 |
Bratzel F., Turck F., 2015. Molecular memories in the regulation of seasonal flowering: from competence to cessation. Genome Biol, 16: 192. DOI:10.1186/s13059-015-0770-6 |
Conti L., 2019. The A-B-A of floral transition: the to do list for perfect escape. Mol.Plant, 12: 289-291. DOI:10.1016/j.molp.2019.02.002 |
de Jong M., Tavares H., Pasam R.K., et al, 2019. Natural variation in Arabidopsis shoot branching plasticity in response to nitrate supply affects fitness. PLoS Genet, 15: e1008366. DOI:10.1371/journal.pgen.1008366 |
Fischer J.J., Beatty P.H., Good A.G., et al, 2013. Manipulation of microRNA expression to improve nitrogen use efficiency. Plant Sci, 210: 70-81. DOI:10.1016/j.plantsci.2013.05.009 |
Fredes I., Moreno S., Diaz F.P., et al, 2019. Nitrate signaling and the control of Arabidopsis growth and development. Curr. Opin. Plant Biol, 47: 112-118. DOI:10.1016/j.pbi.2018.10.004 |
Gras D.E., Vidal E.A., Undurraga S.F., et al, 2018. SMZ/SNZ and gibberellin signaling are required for nitrate-elicited delay of flowering time in Arabidopsis thaliana. J. Exp. Bot, 69: 619-631. DOI:10.1093/jxb/erx423 |
Hu J.Y., Zhou Y., He F., et al, 2014. MIR824-regulated AGAMOUS-LIKE 16 contributes to flowering time repression in Arabidopsis. Plant Cell, 26: 2024-2037. DOI:10.1105/tpc.114.124685 |
Hwang K., Susila H., Nasim Z., et al, 2019. Arabidopsis ABF3 and ABF4 transcription factors act with the NF-YC complex to regulate SOC1 expression and mediate drought-accelerated flowering. Mol. Plant, 12: 489-505. DOI:10.1016/j.molp.2019.01.002 |
Johanson U., West J., Lister C., et al, 2000. Molecular analysis of FRIGIDA, a major determinant of natural variation in arabidopsis flowering time. Science, 290: 344-347. DOI:10.1126/science.290.5490.344 |
Kant S., Peng M., Rothstein S.J., 2011. Genetic regulation by NLA and microRNA827 for maintaining nitrate-dependent phosphate homeostasis in Arabidopsis. PLoS Genet, 7: e1002021. DOI:10.1371/journal.pgen.1002021 |
Klebs G., 1913. Uber das Verhältnis der Außenwelt zur Entwicklung der Pflanze. Sitz-Ber Akad Wiss Heidelberg Ser B, 5: 3-47. |
Konishi M., Yanagisawa S., 2013. Arabidopsis NIN-like transcription factors have a central role in nitrate signalling. Nat. Commun, 4: 1617. DOI:10.1038/ncomms2621 |
Korves T.M., Schmid K.J., Caicedo A.L., et al, 2007. Fitness effects associated with the major flowering time gene FRIGIDA in Arabidopsis thaliana in the field. Am.Nat, 169: E141-E157. DOI:10.1086/513111 |
Li H., Hu B., Chu C.C., 2017. Nitrogen use efficiency in crops: lessons from Arabidopsis and rice. J. Exp. Bot, 68: 2477-2488. DOI:10.1093/jxb/erx101 |
Liang G., He H., Yu D.Q., 2012. Identification of nitrogen starvation-responsive MicroRNAs in Arabidopsis thaliana. PloS One, 7: e48951. DOI:10.1371/journal.pone.0048951 |
Lin Y.L., Tsay Y.F., 2017. Influence of differing nitrate and nitrogen availability on flowering control in Arabidopsis. J. Exp. Bot, 68: 2603-2609. DOI:10.1093/jxb/erx053 |
Liu T., Li Y., Ren J., Qian Y., Yang X., Duan W., Hou X., 2013. Nitrate or NaCl regulates floral induction in Arabidopsis thaliana. Biologia, 68: 215-222. DOI:10.2478/s11756-013-0004-x |
Marin I.C., Loef I., Bartetzko L., et al, 2011. Nitrate regulates floral induction in Arabidopsis, acting independently of light, gibberellin and autonomous pathways. Planta, 233: 539-552. DOI:10.1007/s00425-010-1316-5 |
Michaels S.D., 2009. Flowering time regulation produces much fruit. Curr. Opin.Plant Biol, 12: 75-80. DOI:10.1016/j.pbi.2008.09.005 |
Mishra P., Panigrahi K.C., 2015. Gigantea-an emerging story. Front. Plant Sci, 6: 8. |
Nee G., Xiang Y., Soppe W.J.J., 2017. The release of dormancy, a wake-up call for seeds to germinate. Curr. Opin. Plant Biol, 35: 8-14. DOI:10.1016/j.pbi.2016.09.002 |
Noguero M., Lacombe B., 2016. Transporters involved in root nitrate uptake and sensing by arabidopsis. Front. Plant Sci, 7: 1391. |
Olas J.J., Van Dingenen J., Abel C., Dzialo M.A., Feil R., Krapp A., Schlereth A., Wahl V., 2019. Nitrate acts at the Arabidopsis thaliana shoot apical meristem to regulate flowering time. New Phytol, 223: 814-827. DOI:10.1111/nph.15812 |
Pant B.D., Musialak-Lange M., Nuc P., et al, 2009. Identification of nutrientresponsive Arabidopsis and rapeseed microRNAs by comprehensive real-time polymerase chain reaction profiling and small RNA sequencing. Plant Physiol, 150: 1541-1555. DOI:10.1104/pp.109.139139 |
Pigliucci M., Schlichting C.D., 1995. Reaction norms Arabidopsis (Brassicaceae). Ⅲ.Response to nutrients in 26 populations from a worldwide collection. Am. J. Bot, 82: 1117-1125. DOI:10.1002/j.1537-2197.1995.tb11582.x |
Reeves P.H., Coupland G., 2001. Analysis of flowering time control in Arabidopsis by comparison of double and triple mutants. Plant Physiol, 126: 1085-1091. DOI:10.1104/pp.126.3.1085 |
Riboni M., Galbiati M., Tonelli C., Conti L., 2013. Gigantea enables drought escape response via abscisic acid-dependent activation of the florigens and suppressor of overexpression of constans1. Plant Physiol, 162: 1706-1719. DOI:10.1104/pp.113.217729 |
Riboni M., Test A.R., Galbiati M., 2016. ABA-dependent control of GIGANTEA signalling enables drought escape via up-regulation of FLOWERING LOCUS T in Arabidopsis thaliana. J. Exp. Bot, 67: 6309-6322. DOI:10.1093/jxb/erw384 |
Song Y.H., Estrada D.A., Johnson R.S., et al, 2014. Distinct roles of FKF1, GIGANTEA, and ZEITLUPE proteins in the regulation of CONSTANS stability in Arabidopsis photoperiodic flowering. Proc. Natl. Acad. Sci. Unit. States Am, 111: 17672-17677. DOI:10.1073/pnas.1415375111 |
Srikanth A., Schmid M., 2011. Regulation of flowering time: all roads lead to Rome. Cell. Mol. Life Sci, 68: 2013-2037. DOI:10.1007/s00018-011-0673-y |
Stinchcombe J.R., Weinig C., Ungerer M., et al, 2004. A latitudinal cline in flowering time in Arabidopsis thaliana modulated by the flowering time gene FRIGIDA. Proc. Natl. Acad. Sci. U. S. A, 101: 4712-4717. DOI:10.1073/pnas.0306401101 |
Stitt M., 1999. Nitrate regulation of metabolism and growth. Curr. Opin. Plant Biol, 2: 178-186. DOI:10.1016/S1369-5266(99)80033-8 |
Tschoep H., Gibon Y., Carillo P., et al, 2009. Adjustment of growth and central metabolism to a mild but sustained nitrogen-limitation in Arabidopsis. Plant Cell Environ, 32: 300-318. DOI:10.1111/j.1365-3040.2008.01921.x |
Turck F., Fornara F., Coupland G., 2008. Regulation and identity of florigen: FLOWERING LOCUS T moves center stage. Annu. Rev. Plant Biol, 59: 573-594. DOI:10.1146/annurev.arplant.59.032607.092755 |
Wahl V., Ponnu J., Schlereth A., et al, 2013. Regulation of flowering by trehalose-6-phosphate signaling in Arabidopsis thaliana. Science, 339: 704-707. DOI:10.1126/science.1230406 |
Wang J.W., Czech B., Weigel D., 2009. miR156-regulated SPL transcription factors define an endogenous flowering pathway in Arabidopsis thaliana. Cell, 138: 738-749. DOI:10.1016/j.cell.2009.06.014 |
Wang W., Hu B., Yuan D., et al, 2018. Expression of the nitrate transporter gene OsNRT1. 1A/OsNPF6, 3 confers high yield and early maturation in rice. Plant Cell, 30: 638-651. DOI:10.1105/tpc.17.00809 |
Weber K., Burow M., 2018. Nitrogen-essential macronutrient and signal controlling flowering time. Physiol. Plantarum, 162: 251-260. DOI:10.1111/ppl.12664 |
Wu G., Poethig R.S., 2006. Temporal regulation of shoot development in Arabidopsis thaliana by miR156 and its target SPL3. Development, 133: 3539-3547. DOI:10.1242/dev.02521 |
Wu K., Wang S., Song W., et al, 2020. Enhanced sustainable green revolution yield via nitrogen-responsive chromatin modulation in rice. Science, 367: eaaz2046. DOI:10.1126/science.aaz2046 |
Yuan S., Zhang Z.-W., Zheng C., et al, 2016. Arabidopsis cryptochrome 1 functions in nitrogen regulation of flowering. Proc. Natl. Acad. Sci. U. S. A, 113: 7661-7666. DOI:10.1073/pnas.1602004113 |



