b. University of Chinese Academy of Sciences, Beijing 100049, People's Republic of China
The establishment of a transgenic plant line largely relies on an efficient screening system that can select transformed cells through the expression of a selectable marker gene. The most commonly used selectable markers are the nptII, hpt, and bar genes, which confer antibiotic (kanamycin, hygromycin) or herbicide (phosphinothricin) resistance (Rosellini, 2012). These selectable marker genes have generated great public concern owing to their potentially adverse effects on both human health and the environment (Lucht, 2015). To address this problem, researchers have generated marker-free transgenic plants by removing selectable marker genes by co-transformation, homologous recombination, site-specific recombination, or CRISPR-based targeted degradation systems (Tuteja et al., 2012, Kamthan et al., 2016). However, these approaches are complicated and time consuming. One alternative strategy to address this issue is to use selectable marker genes that are involved in sugar metabolism of most organisms, e.g., phosphomannose isomerase (PMI) (Joersbo et al., 1998), or xylose isomerase (XYL) (Haldrup et al., 1998).
PMI catalyzes the reversible isomerization of mannose-6-phosphate to fructose-6-phosphate. Plant cells generally display too low an activity of PMI to utilize mannose as a sole carbon source. In contrast, bacteria, fungi, and certain algae are able to use mannose to maintain normal growth, and transformed cells expressing PMI can utilize mannose as a carbohydrate source and grow and develop normally in mannose media. PMI isolated from Escherichia coli (Migula) Castellani et Chalmers has been successfully used as a selectable marker gene for the transformation of a number of plant species, including sugar beet, maize, wheat, and rice (Stoykova and Stoeva-Popova, 2010). However, bacterial PMI has raised public safety concerns (Miki and McHugh, 2004). Plant-type PMIs may offer a superior alternative.
Microalgae, which are the primary source of food and nutraceutical consumption for aquatic animals and humans, play an important role in ecosystems and the economy (Ghosh et al., 2016, Sathasivam et al., 2019). Green algae are the ancestors of higher plants (De Vries and Archibald, 2018). In addition to growing photo-autotrophically, some green algae (e.g., Chlorella zofingiensis Donz and Chlorococcum sp.) can grow in the dark using various sugars, including mannose, as a sole carbon source (Sun et al., 2008, Tanoi et al., 2011, Ma and Chen, 2001). Here, we propose using green algae PMI genes as a novel selectable marker for plant biotechnology.
In this study, we characterized PMI from the green algae Chlorococcum sp. and determined its feasibility as a selectable marker gene for genetic transformation of tomato (Solanum lycopersicum L.), an important crop and major source of carotenoids and vitamin C. We found that ChlPMI is more homologous to higher plant PMIs than to bacterial PMIs. Overexpression of ChlPMI in two tomato cultivars resulted in callus and shoot formation in media containing mannose (6 g/L) and had a transformation rate up to 4.5%. The algal PMI gene can be used in tomato genetic engineering and serve as a selectable marker gene for plant biotechnology.
2. Materials and methods 2.1. Isolation and sequence analysis of ChlPMIChlorococcum sp. were isolated from a rocky wall at Victoria Peak, Hong Kong (Ma and Chen, 2001). The alga was cultured following previous protocols (Huang et al., 2012). Total RNA was isolated from aliquots of about 108 cells using the TRI reagent (Molecular research center, Cincinnati, OH, USA) according to the manufacturer's instructions. The concentration of total RNA was determined spectrophotometrically at 260 nm. Reverse transcription was performed using Reverse Transcription System (Omega) according to the manufacturer's instructions. PMI cDNA was isolated by using a pair of degenerate primers designed for the amplification of a partial PMI cDNA, from which specific primers were derived for 3′ and 5′ RACE according to an approach we previously developed (Huang and Chen, 2006). Primers used for cloning cDNAs are listed in Table S1.
Bioinformatic analyses were performed using the BioEdit program and NCBI online tools. The 3D structure was predicted by SWISS-MODEL (https://swissmodel.expasy.org/).
2.2. Phylogenetic analysisTo compare the evolutionary relationships between PMI proteins, a homology search was performed by BLASTP (http://blast.ncbi.nlm.nih.gov/Blast.cgi) using the amino acid sequence of ChlPMI. Amino acid sequences of PMIs (from 6 prokaryotic and 10 eukaryotic organisms) were downloaded from NCBI and were used for phylogenetic analysis. Multiple sequence alignments were conducted with the ClustalW program of MEGA7 software, and a tree was constructed with MEGA7 software using the Neighbor-Joining method (Kumar et al., 2016). The parameters were Poisson model, pairwise deletion, and bootstrap analysis with 1000 replicates.
2.3. Effect of mannose on organogenesis of tomato cotyledon explantsTo evaluate the effect of mannose on the organogenesis, Micro-Tom tomato cotyledons were plated on MS media with various concentrations of a mixture of mannose and sucrose (Table 1). For each combination, 8 petri dishes were used. The dishes were cultured under a 16-h light/8-h dark photoperiod at 25 ℃. The number of explants with callus tissue was determined after 6 weeks of cultivation, and the number of explants with shoots was measured after 8 weeks. Regeneration rate was calculated as the ratio of the number of explants with callus or shoots to total number of plated explants.
| Medium | Mannose (g/L) | Sucrose (g/L) | Number of explants | Explants with callus number | Explants with shoot number | Callus/shoot Regeneration rate (%) |
| a | 0 | 20 | 201 | 185 | 124 | 92/61 |
| b | 3 | 17 | 205 | 112 | 31 | 55/15 |
| c | 6 | 14 | 207 | 7 | 0 | 3/0 |
| d | 9 | 11 | 204 | 0 | 0 | 0/0 |
The ORF of ChlPMI was cloned downstream of the NOS promoter to replace the nptII in pBI121, resulting in the vector pBI-PMI (Supplement Fig. 1a). In addition, a polycistronic gene cluster linked by 2A was inserted into pBI-PMI to replace the GUS gene, leading to pBI-PMI-BBBB (Supplement Fig. 1b). pBI-PMI-BBBB confers enhanced carotenoid production to plant cells via Erwinia uredovora (Pon et al.) Dye crtB (GenBank: BAA14128) and S. lycopersicum LCYB (GenBank: 544104). In addition, the vector confers the ability to synthesize red ketocarotenoids via Haematococcus pluvialis Flotow emend. Wille BHY (GenBank: Q9SPK6), Chlamydomonas reinhardtii P.A. Dangeard BKT (GenBank: AEA35045).
|
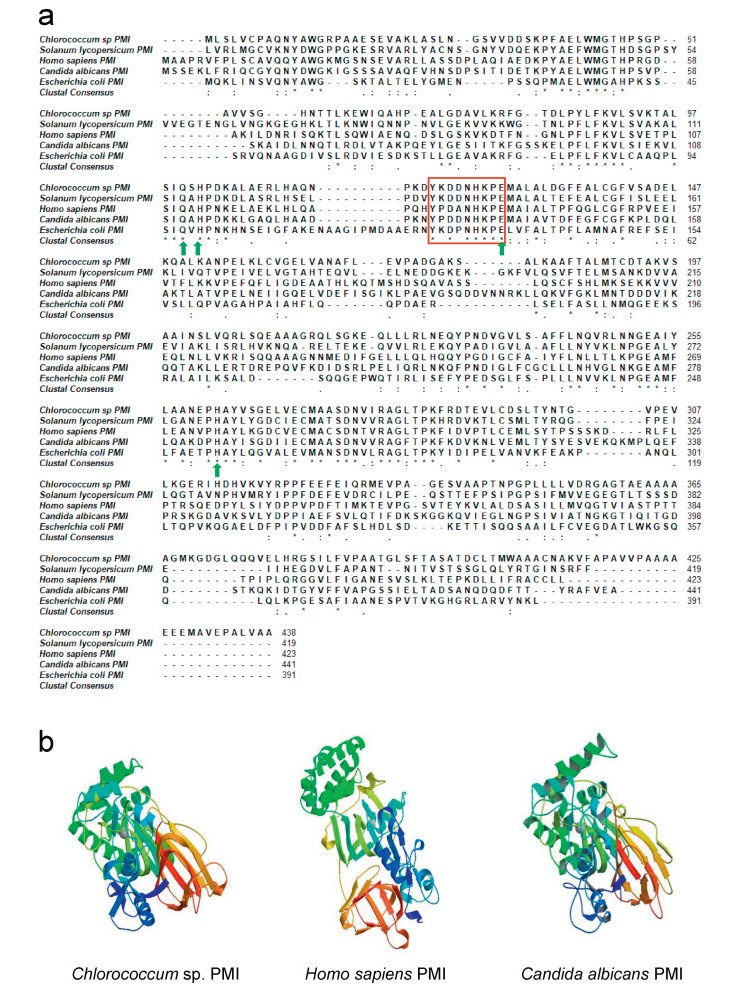
| Fig. 1 Sequence alignment (a) and structures (b) of classic type Ⅰ PMIs from Chlorococcum sp. and other species. The conserved YXDXNHKPE motif is boxed in red. Green arrows label four residues (Gln-111, Glu-138, His-113 and His-285). Accession numbers of the PMI: Solanum lycopersicum-XP_004233489.1; Homo sapiens-CAA53657.1; Candida albicans-CAA57548.1; Escherichia coli- NP_416130.3. |
Wild-type tomato seeds of Micro-Tom and Beta mutant LA0316 were kept by our laboratory. Cotyledons cut from 14-day-old seedlings were used for transformation following the methods described previously (Huang et al., 2013) except for the use of 6 g/L mannose as the selective agent. The mannose-resistant plants were planted in a greenhouse and used for further analysis.
2.5. Polymerase chain reaction (PCR) analysis of mannose-resistant plantsGenomic DNA was isolated from leaves using the CTAB method. Specific primers (listed in Table S1) were designed to detect the existence of ChlPMI, crtB, HpBHY, and CrBKT in regenerated plants selected by mannose-containing media. PCR reactions were carried out using 2×Taq PCR Mix (Tsingke, Kunming, China) with the program: 2 min at 94 ℃ followed by 32 cycles of 94 ℃ for 20 s, 60 ℃ for 20 s, 72 ℃ for 30 s, and a final extension of 5 min 72 ℃. The resulting products were analyzed by electrophoresis on a 1.5% agarose gel.
2.6. Chlorophenol red assayPMI enzyme activity was detected with the chlorophenol red assay according to Guo et al. (2015). Transgenic and wild-type leaf tissues (approximately 0.4 × 0.6 cm2), were incubated in the dark at 27 ℃ in MS liquid medium (pH 6.0) with 6 g/L mannose and 50 mg/L chlorophenol red pH indicator, and then assessed for color changes after 4 days. Changes in medium color indicate the ability of transgenic plants to metabolize mannose.
2.7. Pigment extraction and analysisPigments were extracted and analyzed as described in our previous studies (Huang et al., 2013, Ye et al., 2019). Pigments were identified and calculated through comparison with standard curves of authentic pigments. Data are shown as means ± SD (Standard Deviation) and the significant differences between PMI-BBBB and WT were examined by Student's t-test.
3. Results 3.1. Isolation and molecular characterization of Chlorococcum sp. PMI (ChlPMI)Using the RACE technique, we isolated ChlPMI cDNA that has an ORF of 1314 bp (deposited in GenBank, Accession number: MT267358) and encodes a polypeptide of 438 amino acids (molecular mass = 46.42 kDa). The isolated ChlPMI belongs to the classic PMI type Ⅰ family. Sequence alignments showed that ChlPMI contains the PMI consensus sequence YXDXNHKPE (Fig. 1a) and four conserved amino acids (Gln-111, Glu-138, His-113 and His-285) that form the zinc-binding site and therefore play a vital role in enzymatic activity (Cleasby et al., 1996). In addition, the three-dimensional (3D) structure of ChlPMI exhibits high similarity to that of other classic type Ⅰ PMIs (Fig. 1b).
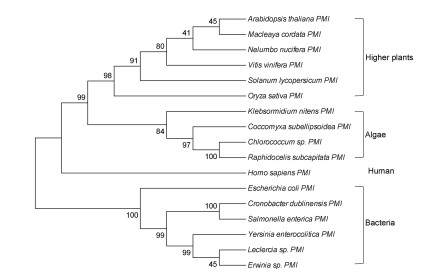
A phylogenetic tree was constructed using the amino acid sequences of ChlPMI and 16 other PMIs from 6 bacteria, 3 algae, 6 higher plant species and human. Phylogenetic analysis revealed that ChlPMI is clustered in the subgroup with algal PMIs and is a member of a large branch with plant PMIs and human PMI. In contrast, ChlPMI was distant to bacterial PMIs which were branched-off separately (Fig. 2).
|
| Fig. 2 Phylogenetic tree of type Ⅰ PMIs. MEGA7 software was used to construct the tree using the Neighbor-Joining method with 1000 bootstrap replicates. The bootstrap support values showing the confidence level are given at the clade nodes as percentages. |
Tomato is an important crop in which much transgenic research has been performed (Bergougnoux, 2014). Thus, we aimed to determine whether ChlPMI could serve as an effective selectable marker gene for genetic manipulation of tomato. To this end, we first examined organogenesis of tomato cotyledon explants (from the tomato cultivar Micro-Tom) grown on MS media exposed to various concentrations of mannose and sucrose as carbon sources (Table 1). Of the 201 tomato explants grown on medium with no mannose, 185 (92%) produced callus tissue and 124 (61%) produced shoots (Table 1). In contrast, when the medium contained 3 g/L mannose, the callus and shoot induction rates decreased to 55% and 15%. However, when the medium consisted of mannose up to 6 g/L, no shoots were induced (Table 1 and Supplement Fig. 2). This result confirmed that tomato tissue was sensitive to mannose, possibly because of low endogenous PMI activity. Thus, we next investigated whether the heterologous expression of ChlPMI allowed tomato cells to grow on media containing mannose up to 6 g/L.
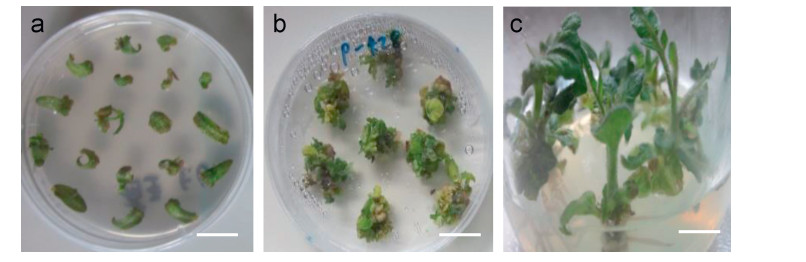
3.3. Overexpression of ChlPMI in tomatoTo assess whether ChlPMI could act as an effective selectable marker for plant transformation, ChlPMI was cloned downstream of the NOS promoter in the pBI121 binary vector to replace nptII, resulting in the vector pBI-PMI (Supplement Fig. 1a). pBI-PMI was introduced into the tomato Micro-Tom cultivar via Agrobacterium-mediated transformation (Huang et al., 2013); mannose rather than kanamycin was used as a selective agent. Three independent experiments were performed in which a total of 428 cotyledon explants were transformed. The results are summarized in Table 2. Unsuccessfully transformed cotyledon explants were prone to browning and necrosing after two months of mannose selection (Fig. 3a); in contrast, transformed cells formed callus tissue, from which buds and shoots were gradually generated (Fig. 3b). Normal plantlets were produced after the shoots were rooted in a mannose-containing rooting medium (Fig. 3c).
| Experiment | Number of explants | Explants with callus number | Number of shoots regenerated from callus | Number of seedlings | PCR positive plants | Transformation rate (%) |
| 1 | 180 | 79 | 51 | 7 | 6 | 3.3% |
| 2 | 170 | 83 | 68 | 7 | 7 | 4.1% |
| 3 | 178 | 81 | 60 | 9 | 8 | 4.5% |
|
| Fig. 3 Regeneration of transformed Micro-Tom tomato expressing ChlPMI on MS medium with 6 g/L mannose. (a) Untransformed cotyledon explants. (b) Shoots and callus tissues generated from transformed cells. (c) Regenerated seedlings. Bar = 1.5 cm. |
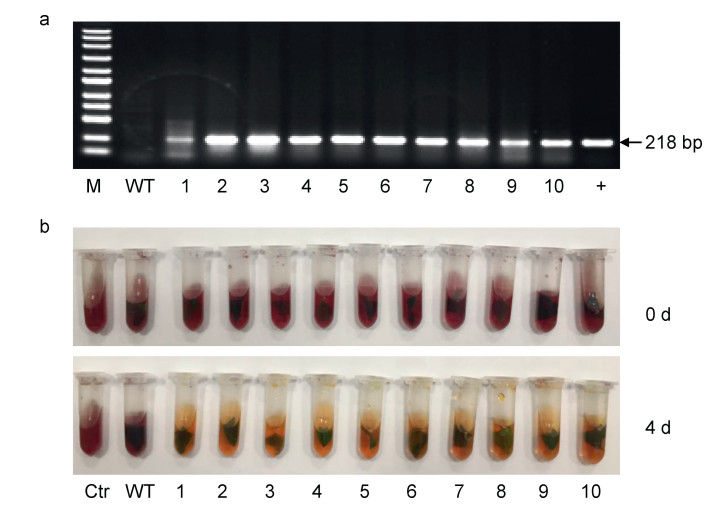
Normal seedlings generated from rooting medium supplemented with 6 g/L mannose were considered putative transgenic lines. To confirm that these seedlings were transgenic, we amplified a 218-bp fragment from ChlPMI gene. PCR amplification showed that out of 23 lines obtained from three batches of experiments, 21 were transgenic plants (Fig. 4a). Thus, the average transformation rate (expressed as the ratio of ChlPMI-expressing plants to total number of plated explants) was 3.9%.
|
| Fig. 4 PCR detection (a) and chlorophenol red assay (b) of regenerated tomato plants. M-100 bp DNA Ladder; WT-Micro-Tom cultivar; 1–10-regenerated lines; +-pBI-PMI plasmid. Ctr-negative control without leaf tissue. |
To further confirm the expression of ChlPMI in the putative transgenic plants, we performed a chlorophenol red assay that evaluates PMI activity in plant tissues (Guo et al., 2015). Leaf segments of transgenic plants confirmed by PCR detection and WT control plants were incubated in MS liquid medium supplemented with chlorophenol red and mannose. The pH value of the medium was initially 6.0, as indicated by the purple color of the liquid. Visual differences were observed after four days of incubation; all media containing leaves of transformed plants turned yellow and brown, whereas media with WT leaves remained purple (Fig. 4b). The colorimetric change indicated that the transformed plants had acidified the medium, and were thus able to utilize mannose, indicating that heterologous PMI is enzymatically active in the transgenic plants.
The putative transgenic plantlets were planted in a greenhouse. We found that the transformants grew normally with a phenotype consistent with WT plants (Supplement Fig. 3). Thus, using ChlPMI as a selectable marker, we established a reliable and efficient ChlPMI/mannose system for tomato transformation.
3.4. Astaxanthin-producing tomato generated by the ChlPMI/mannose systemWe previously generated astaxanthin-producing transgenic tomato lines using the traditional kanamycin selection system (Huang et al., 2013). However, the kanamycin selectable marker gene hinders commercial application of nutrient-rich tomato lines. Hence, we attempted to generate astaxanthin-producing tomato lines using our ChlPMI/mannose system. We co-expressed ChlPMI and a polycistronic cassette containing four rate-limiting carotenogenic genes that encode phytoene synthase (PSY/crtB), β-carotene hydroxylase (BHY), β-carotene ketolase (BKT), and Lycopene β-cyclase (LCYB) in the tomato Beta cultivar. The polycistronic cassette confers the ability to synthesize the red ketocarotenoid astaxanthin to plant cells, which changes the color of callus tissue and regenerated shoots from green in WT to brown in transgenic tissues. As a result, transgenic lines can be easily identified.
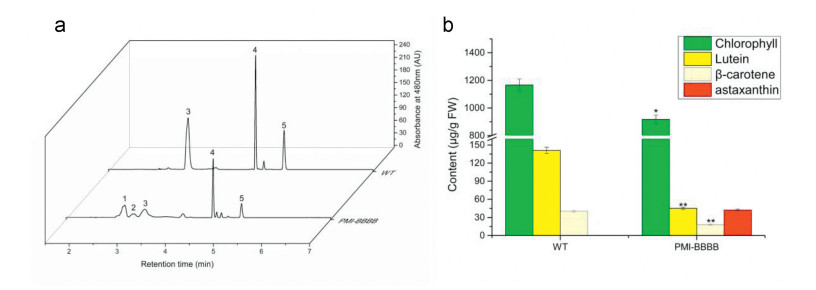
Six independent lines of putative transgenic tomato were generated (Table S2). PCR detection showed the presence of transgenes in all the six lines (Supplement Fig. 4). Before planted in soil, transgenic plantlets shared similar characteristics with WT, except for the color (Fig. 5a). However, after planted into soil, transgenic plants grew much slower than WT plants, and the young leaves of the transgenic plants, especially near the stem apex and axillary bud, were shriveled and bleached. Without blossoming and fruiting, all transgenic lines eventually died (Fig. 5b and c). This may have been the result of overexpression of carotenogenic genes, which have been demonstrated to lead to interference of endogenous metabolic networks and normal plant development (Fray et al., 1995). For example, clear evidence of pigment biosynthesis was exhibited in transgenic plants, which were brownish red rather than green. We extracted and analyzed the pigments in leaves of transgenic and WT seedlings using Ultra-performance liquid chromatography (UPLC). UPLC analysis showed that besides common pigments (chlorophylls, lutein, and β-carotene), transgenic lines produced two new pigments, astaxanthin and ketolutein (Fig. 6a). The accumulation of ketocarotenoids led to the sharp decrease of the primary pigments, including chlorophylls, lutein and β-carotene (Fig. 6b), which might impact the photosynthesis of the transgenic lines and hence plant growth.

|
| Fig. 5 Growth status of WT and PMI-BBBB plants. Plantlets of WT (left) and PMI-BBBB (right) (a) were planted in a greenhouse at flowering (b), and fruiting (c). WT-Beta cultivar. Bar = 1.5 cm. |
|
| Fig. 6 Pigment analysis in leaves of WT and PMI-BBBB. UPLC analysis of pigments (a) and contents of main pigments (b) in tomato leaves of WT and PMI-BBBB. WT-Beta cultivar. Identified pigments of peaks: 1-astaxanthin; 2-ketolutein; 3-lutein; 4-chlorophyll; 5-β-carotene. Data represent average values from measurements of three individual tomato leaves. Significant differences were determined by Student's t-tests. The P-Values are indicated as follows: *P < 0.05, **P < 0.01. |
In this study, we determined the feasibility of using a green algal PMI gene as a selectable marker to create transgenic plants. PMIs, which are biologically safer selectable markers than nptII, hpt and bar (Breyer et al., 2014, Miki and McHugh, 2004), have been used to genetically modify Golden Rice (Oryza sativa L.) (Hoa et al., 2003) and maize (Zea mays L.) (European Food Safety Authority, 2009), in accordance to regulatory requirements. The bacterial PMI has been applied in plant transformation (Stoykova and Stoeva-Popova, 2010). However, although no evidence suggests that consuming foods genetically modified with bacterial PMI is harmful to human health or the environment, consumers are averse to such genetically modified crops (Reed et al., 2001, Privalle, 2002). Previous studies have reasoned that, in accordance with the principles of cisgenesis and intragenesis (Holme et al., 2013), using genes from species phylogenetically related to plants might facilitate social acceptance of genetically modified crops (Lucht, 2015). Therefore, given the plant-like characteristics of algae, we speculated that green alga rather than bacterial PMI would be a superior selectable marker.
Certain algae utilize mannose as a sole carbon source for heterotrophic growth (Sun et al., 2008, Tanoi et al., 2011, Ma and Chen, 2001), suggesting algae PMI genes can be used as selectable markers for plant manipulation. Here, we characterized the PMI gene from the edible algae Chlorococcum sp., which encodes a putative classic type Ⅰ phosphomannose isomerase with all the conserved domains and motifs of typical PMIs (Fig. 1a). Phylogenetic analysis showed that ChlPMI clustered together with plant-type PMIs and human PMI but was distant to bacterial PMIs (Fig. 2).
Although PMI genes are universal in the genomes of both prokaryotic and eukaryotic organisms, many higher plants, such as Arabidopsis (Maruta et al., 2008), Chinese cabbage (Wang et al., 2014) and O. sativa (Hu et al., 2016) are sensitive to mannose, possibly due to no or low PMI expression. Our study showed that tomato is also sensitive to mannose. Hence, when tomato cotyledon explants were cultured in medium containing mannose (6 g/L), no shoots were induced. In contrast, shoots were regenerated from explants on the same culture medium when ChlPMI was expressed in tomato cells (Fig. 3b). PMI activity was detected in the transgenic plants (Fig. 4b), but the heterologous expression of ChlPMI did not interfere with the growth and development of tomato (Supplement Fig. 3), confirming that PMI is a "phenotypically neutral" selectable marker (Rosellini, 2012). The transformation ratio reached up to 4.5% (Table 2). Previously, different transformation ratios (2.0–15.5%) were achieved in two different tomato cultivars expressing a bacterial PMI, depending on the construct, genotype, and type of tissue used for transformation (Sigareva et al., 2004, Briza et al., 2008). Similarly, transformation efficiency in Citrus sinensis (L.) Osbeck ranges from 1.6% to 23.8% in different cultivars (Boscariol et al., 2003, Wu et al., 2019). Thus, when PMI is used as a selectable marker, genotypes and explant type should be coupled with optimal selection medium.
Major goals of tomato breeding include improving carotenoid profiles in tomato fruit (Liu et al., 2015, Rodriguez-Concepcion et al., 2018), yield, stress resistance, and other agronomic traits. A PMI/mannose-based selection system may play an important role achieving these goals. In the present study, we demonstrated that ChlPMI is an efficient selectable marker gene for tomato engineering. Furthermore, we successfully generated astaxanthin-producing tomato based on this selection system. The engineered tomato (PMI-BBBB) synthesized astaxanthin in leaves (Fig. 5, Fig. 6). The low transformation rate (1.4%) for astaxanthin-producing tomato could result from (1) the large expression cassette; (2) Beta mutants being less sensitive to mannose; and/or (3) astaxanthin alleviating shoot induction, which was supported by the formation of different color (red and green) callus tissues and shoots. Thus, to elevate the transformation efficiency, the screening system needs optimization. Furthermore, the activity of ChlPMI can be increased by codon optimization and/or altering the promoter and terminator as done by previous studies (Sigareva et al., 2004, Briza et al., 2008, Gui et al., 2014).
Author contributionsY. Y. L. performed the experiments and wrote the manuscript. J. C. H. designed the experiments and wrote the manuscript. The authors have read and approved the manuscript.
Declaration of Competing InterestThe authors declare that they have no conflict of interest.
AcknowledgmentsThe work described in this paper was supported by a grant from Yunnan high talents program (Y33D331), Yunnan Province, China.
Appendix A. Supplementary dataSupplementary data to this article can be found online at https://doi.org/10.1016/j.pld.2020.06.001.
Bergougnoux V., 2014. The history of tomato: from domestication to biopharming. Biotechnol. Adv, 32: 170-189. DOI:10.1016/j.biotechadv.2013.11.003 |
Boscariol R.L., Almeida W.A.B., Derbyshire M., et al, 2003. The use of the PMI/mannose selection system to recover transgenic sweet orange plants (Citrus sinensis L. Osbeck). Plant Cell Rep, 22: 122-128. DOI:10.1007/s00299-003-0654-1 |
Breyer D., Kopertekh L., Reheul D., 2014. Alternatives to antibiotic resistance marker genes for in vitro selection of genetically modified plants-scientific developments, current use, operational access and biosafety considerations. Crit. Rev. Plant Sci, 33: 286-330. DOI:10.1080/07352689.2013.870422 |
Briza J., Pavingerova D., Prikrylova P., et al, 2008. Use of phosphomannose isomerase-based selection system for Agrobacterium-mediated transformation of tomato and potato. Biol. Plant. (Prague), 52: 453-461. DOI:10.1007/s10535-008-0090-8 |
Cleasby A., Wonacott A., Skarzynski T., et al, 1996. The X-ray crystal structure of phosphomannose isomerase from Candida albicans at 1. 7 angstrom resolution. Nat. Struct. Biol, 3: 470-479. DOI:10.1038/nsb0596-470 |
De Vries J., Archibald J.M., 2018. Plant evolution: landmarks on the path to terrestrial life. New Phytol, 217: 1428-1434. DOI:10.1111/nph.14975 |
European Food Safety Authority (EFSA), 2009. Application (Reference EFSA-GMOUK-2005-11) for the placing on the market of insect-resistant genetically modified maize MIR604 event, for food and feed uses, import and processing under Regulation (EC) No 1829/2003 from Syngenta Seeds S.A. S on behalf of Syngenta Crop Protection AG. EFSA J 1193, 1-26.
|
Fray R.G., Wallace A., Fraser P.D., et al, 1995. Constitutive expression of a fruit phytoene synthase gene in transgenic tomatoes causes dwarfism by redirecting metabolites from the gibberellin pathway. Plant J, 8: 693-701. DOI:10.1046/j.1365-313X.1995.08050693.x |
Ghosh A., Khanra S., Mondal M., et al, 2016. Progress toward isolation of strains and genetically engineered strains of microalgae for production of biofuel and other value added chemicals: a review. Energy Convers. Manag, 113: 104-118. DOI:10.1016/j.enconman.2016.01.050 |
Gui H., Li X., Liu Y., et al, 2014. The relationship between PMI (manA) gene expression and optimal selection pressure in Indica rice transformation. Plant Cell Rep, 33: 1081-1090. DOI:10.1007/s00299-014-1596-5 |
Guo Q., Ma J., Yuan B., et al, 2015. High-efficiency Agrobacterium-mediated transformation of Lotus corniculatus L. using phosphomannose isomerase positive selection. Plant Cell Tissue Organ, 121: 413-422. DOI:10.1007/s11240-015-0712-8 |
Haldrup A., Petersen S.G., Okkels F.T., 1998. Positive selection: a plant selection principle based on xylose isomerase, an enzyme used in the food industry. Plant Cell Rep, 18: 76-81. DOI:10.1007/s002990050535 |
Hoa T.T., Al-Babili S., Schaub P., et al, 2003. Golden Indica and Japonica rice lines amenable to deregulation. Plant Physiol, 133: 161-169. DOI:10.1104/pp.103.023457 |
Holme I.B., Wendt T., Holm P.B., 2013. Intragenesis and cisgenesis as alternatives to transgenic crop development. Plant Biotechnol. J, 11: 395-407. DOI:10.1111/pbi.12055 |
Hu L., Li H., Qin R., et al, 2016. Plant phosphomannose isomerase as a selectable marker for rice transformation. Sci. Rep, 6: 25921. DOI:10.1038/srep25921 |
Huang J., Zhong Y., Sandmann G., et al, 2012. Cloning and selection of carotenoid ketolase genes for the engineering of high-yield astaxanthin in plants. Planta, 236: 691-699. DOI:10.1007/s00425-012-1654-6 |
Huang J.C., Chen F., 2006. Simultaneous amplification of 5' and 3' cDNA ends based on template-switching effect and inverse PCR. Biotechniques, 40: 187-189. DOI:10.2144/000112051 |
Huang J.C., Zhong Y.J., Liu J., et al, 2013. Metabolic engineering of tomato for highyield production of astaxanthin. Metab. Eng, 17: 59-67. DOI:10.1016/j.ymben.2013.02.005 |
Joersbo M., Donaldson I., Kreiberg J., et al, 1998. Analysis of mannose selection used for transformation of sugar beet. Mol. Breed, 4: 111-117. DOI:10.1023/A:1009633809610 |
Kamthan A., Chaudhuri A., Kamthan M., et al, 2016. Genetically modified (GM)crops: milestones and new advances in crop improvement. Theor. Appl. Genet, 129: 1639-1655. DOI:10.1007/s00122-016-2747-6 |
Kumar S., Stecher G., Tamura K., 2016. MEGA7:molecular evolutionary genetics analysis version 7. 0 for bigger datasets. Mol. Biol. Evol, 33: 1870-1874. DOI:10.1093/molbev/msw054 |
Liu L., Shao Z., Zhang M., et al, 2015. Regulation of carotenoid metabolism in tomato. Mol. Plant, 8: 28-39. DOI:10.1016/j.molp.2014.11.006 |
Lucht J.M., 2015. Public acceptance of plant biotechnology and GM crops. Viruses, 7: 4254-4281. DOI:10.3390/v7082819 |
Ma R.Y.N., Chen F., 2001. Enhanced production of free trans-astaxanthin by oxidative stress in the cultures of the green microalga Chlorococcum sp. Process Biochem, 36: 1175-1179. DOI:10.1016/S0032-9592(01)00157-1 |
Maruta T., Yonemitsu M., Yabuta Y., et al, 2008. Arabidopsis phosphomannose isomerase 1, but not phosphomannose isomerase 2, is essential for ascorbic acid biosynthesis. J. Biol. Chem, 283: 28842-28851. DOI:10.1074/jbc.M805538200 |
Miki B., McHugh S., 2004. Selectable marker genes in transgenic plants: applications, alternatives and biosafety. J. Biotechnol, 107: 193-232. DOI:10.1016/j.jbiotec.2003.10.011 |
Privalle, L.S., 2002. Phosphomannose isomerase, a novel plant selection system-potential allergenicity assessment. In: Fu, T.J., Gendel, S.M. (Eds. ), Genetically Engineered Foods Assessing Potential Allergenicity. New York Acad Sciences, New York, pp. 129-138.
|
Reed J., Privalle L., Powell M.L., et al, 2001. Phosphomannose isomerase: an efficient selectable marker for plant transformation. In Vitro Cell. Dev. Biol.-Plant, 37: 127-132. DOI:10.1007/s11627-001-0024-z |
Rodriguez-Concepcion M., Avalos J., Bonet M.L., et al, 2018. A global perspective on carotenoids: metabolism, biotechnology, and benefits for nutrition and health. Prog. Lipid Res, 70: 62-93. DOI:10.1016/j.plipres.2018.04.004 |
Rosellini D., 2012. Selectable markers and reporter genes: a well furnished toolbox for plant science and genetic engineering. Crit. Rev. Plant Sci, 31: 401-453. DOI:10.1080/07352689.2012.683373 |
Sathasivam R., Radhakrishnan R., Hashem A., et al, 2019. Microalgae metabolites: a rich source for food and medicine. Saudi J. Biol. Sci, 26: 709-722. DOI:10.1016/j.sjbs.2017.11.003 |
Sigareva M., Spivey R., Willits M.G., et al, 2004. An efficient mannose selection protocol for tomato that has no adverse effect on the ploidy level of transgenic plants. Plant Cell Rep, 23: 236-245. DOI:10.1007/s00299-004-0809-8 |
Stoykova P., Stoeva-Popova P., 2010. PMI (manA) as a nonantibiotic selectable marker gene in plant biotechnology. Plant Cell Tissue Organ, 105: 141-148. |
Sun N., Wang Y., Li Y.T., et al, 2008. Sugar-based growth, astaxanthin accumulation and carotenogenic transcription of heterotrophic Chlorella zofingiensis(Chlorophyta). Process Biochem, 43: 1288-1292. DOI:10.1016/j.procbio.2008.07.014 |
Tanoi T., Kawachi M., Watanabe M.M., 2011. Effects of carbon source on growth and morphology of Botryococcus braunii. J. Appl. Phycol, 23: 25-33. DOI:10.1007/s10811-010-9528-4 |
Tuteja N., Verma S., Sahoo R.K., et al, 2012. Recent advances in development of marker-free transgenic plants: regulation and biosafety concern. J. Biosci, 37: 167-197. DOI:10.1007/s12038-012-9187-5 |
Wang X.H., Zhang S., Hu D., et al, 2014. BcPMI2, isolated from non-heading Chinese cabbage encoding phosphomannose isomerase, improves stress tolerance in transgenic tobacco. Mol. Biol. Rep, 41: 2207-2216. DOI:10.1007/s11033-014-3072-2 |
Wu H., Acanda Y., Canton M., et al, 2019. Efficient biolistic transformation of immature Citrus rootstocks using phosphomannose-isomerase selection. Plants, 8: 390. DOI:10.3390/plants8100390 |
Ye J., Liu M., He M., et al, 2019. Illustrating and enhancing the biosynthesis of astaxanthin and docosahexaenoic acid in aurantiochytrium sp. SK4. Mar. Drugs: 17. |



