b. Laboratory of Systematic & Evolutionary Botany and Biodiversity, College of Life Sciences, Zhejiang University, Hangzhou, 310058, China;
c. College of Life and Environmental Sciences, Wenzhou University, Wenzhou, 325035, China
Celtis L. is a genus of 60–70 species of trees, or rarely shrubs, commonly known as hackberries or nettle trees, widespread in warm temperate regions of the Northern Hemisphere and parts of central Africa and South America (Keerler, 1902; Hwang et al., 2003; Martins et al., 2015). Celtis species are grown as ornamental plants, used in traditional medicines (Kim et al., 2005), provide important wildlife habitat and winter food to birds and mammals (Correll and Johnston, 1970), and serve as pollen sources (Krajicek and Williams, 1990). Despite their high economic and ecological importance, the genomic resources necessary for examining the phylogenetic relationships of Celtis are currently unavailable.
Plants in Celtis show a great diversity of growth forms, along with significant divergence in physiological characters caused by strong ecological differences (Whittemore, 2005). Furthermore, there are apparent intergradations between species, which many scholars have deduced are likely a result of interspecific hybridization (Fernald, 1950; Whittemore and Townsend, 2007). Consequently, the genus Celtis has long been known for its taxonomic difficulty and is one of the most poorly understood plant groups (Hooker, 1885; Whittemore, 2005; Sattarian, 2006; Lee et al., 2011). The genus was previously put in either the elm family (Ulmaceae) or a separate family (Celtidaceae). However, the Angiosperm Phylogeny Group Ⅲ (APG Ⅲ) system transferred it into an expanded hemp family (Cannabaceae) (Angiosperm Phylogeny Group Ⅲ, 2009). Subsequent phylogenetic studies have confirmed that Celtis is a member of Cannabaceae and closely related to Trema Lour., Parasponia Miq., Chaetachme Planch., Pteroceltis Maxim., Humulus L. and Cannabis L. (Yang et al., 2013; Zhang et al., 2018). To date, few studies have analyzed the phylogenetic relationships within Celtis. A molecular phylogeny of Cannabaceae based on four plastid loci included six Celtis accessions, which formed two sister clades, with C. madagascariensis Sattarian, C. iguanaea (Jacq.) Sarg. and C. ehrenbergiana (Klotzsch) Liebm. 2 grouped into one clade, and C. biondii Pamp., C. tournefortii Lam., C. sinensis Pers. and C. ehrenbergiana 1 clustered into the other (Yang et al., 2013). Chen et al. (2017) reported a new species (C. neglecta Zi L. Chen & X. F. Jin) from Zhejiang province, and revealed a topology of ((C. neglecta, C. julianae C.K.Schneid.), C. vandervoetiana C.K.Schneid.) with moderate support based on the atpB-rbcL and trnL-F sequences. Clearly, further phylogenetic analysis is required to improve our understanding of the phylogeny and evolution of Celtis.
Traditionally plant lineages have been inferred using plastid introns and inter-genic spacer regions with highly variable loci (Baldauf et al., 2000; Moncalvo et al., 2002). More recently, whole plastomes have increasingly been used for phylogenetic analyses (Williams et al., 2016; Liu et al., 2017; Gu et al., 2019). Studies have shown that whole plastomes are effective when resolving recalcitrant phylogenetic relationships at shallow taxonomic levels (Givnish et al., 2015; Duvall et al., 2016) and at deeper levels (Jansen et al., 2007; Moore et al., 2010). However, the uniparental inheritance of plastomes means that they are not ideal for identifying phylogenetic relationships of species that have undergone interspecific hybridization (Gruenstaeudl et al., 2013; Johnson et al., 2019).
Additional molecular resources can be obtained by genome skimming, which is one of the genome-level sequencing methods first identified by Straub et al. (2012). The high-copy fraction of a genome (such as plastome, mitogenome and nuclear repetitive elements) is sequenced through sampling of low-coverage genome data, and partial sequences with low-copy nuclear loci recovered from genome skimming are provided to design PCR primers or probes for amplification (Cronn et al., 2012; Straub et al., 2012). Moreover, the internal and external transcribed spacers (ITS and ETS), as well as polymorphic nuclear simple sequence repeats (nSSRs) and plastid genomic hotspots are commonly obtained using this technique (Liu et al., 2018, 2020). Genome skimming has been widely used at different taxonomic levels, for intraspecific 'ultra-barcoding' (Kane et al., 2012), intergeneric (Mcpherson et al., 2013; Li et al., 2017; Liu et al., 2018) or phylogenomic studies (Besnard et al., 2013; Malé et al., 2015; Xu et al., 2018; Yu et al., 2018).
In this study, our aim was threefold. First, we aimed to reveal patterns of plastome evolution in Celtis. For this purpose, we used genome skimming sequencing to reconstruct the plastomes of five Celtis species widely distributed in East Asia. We then examined whether plastome data effectively resolves phylogenetic relationships within Celtis. Finally, we developed universal genomic resources (e.g., plastid hotspot regions, polymorphic nuclear SSRs, single or low copy genes) for the genus. Our results provide powerful tools for future studies on the phylogeny, plastome evolution, and population genetics of Celtis.
2. Materials and methods 2.1. Plant materialsFive Celtis species (C. biondii, C. cerasifera C.K.Schneid., C. julianae, C. koraiensis Nakai and C. sinensis) were sampled at Jinming Campus of Henan University (114°18′27.96″E, 34°49′17.59″N), Kaifeng, Henan, China. Fresh leaves of each species were collected for genomic DNA extraction using modified CTAB reagents (Plant DNAzol, Shanghai, China) according to the manufacturer's protocol. Voucher specimens for the five species were deposited at the Herbarium of Henan University (Table 1). For comparative analysis of plastomes, genomic resources development and phylogenetic inference, we downloaded publicly available plastomes of two additional Celtis species (C. biondii and C. tetrandra) (Zhang et al., 2018; Wang et al., 2019) from GenBank database.
| C. biondii | C. cerasifera | C. julianae | C. koraiensis | C. sinensis | |
| Voucher specimen number | LLX18060301 | LLX18060504 | LLX18060506 | LLX18060702 | LLX060703 |
| Total cpDNA size (bp) | 158, 989 | 159, 063 | 159, 046 | 159, 082 | 159, 074 |
| Length of large single-copy (LSC) region | 86, 075 | 86, 089 | 86, 126 | 86, 171 | 86, 163 |
| Length of inverted repeat (IR) region | 26, 891 | 26, 892 | 26, 891 | 26, 891 | 26, 891 |
| Length of small single-copy (SSC) region | 19, 132 | 19, 190 | 19, 138 | 19, 129 | 19, 129 |
| Coding size (bp) | 91, 441 | 91, 450 | 91, 432 | 91, 444 | 91, 444 |
| Intron size (bp) | 22, 008 | 22, 019 | 22, 004 | 22, 005 | 22, 006 |
| Spacer size (bp) | 45, 540 | 45, 594 | 45, 610 | 45, 633 | 45, 624 |
| Total GC content (%) | 36.30 | 36.30 | 36.30 | 36.30 | 36.30 |
| GC content in LSC region (%) | 34.00 | 34.10 | 34.00 | 34.00 | 34.00 |
| GC content in IR region (%) | 42.30 | 42.30 | 42.30 | 42.30 | 42.30 |
| GC content in SSC region (%) | 29.80 | 29.90 | 29.90 | 29.90 | 29.90 |
| Total number of genes | 112 | 112 | 112 | 112 | 112 |
| Protein encoding | 78 | 78 | 78 | 78 | 78 |
| tRNA | 30 | 30 | 30 | 30 | 30 |
| rRNA | 4 | 4 | 4 | 4 | 4 |
| Number of genes duplicated in IR | 20 | 20 | 20 | 20 | 20 |
| GenBank accession number | MN599037 | MN640571 | MN599038 | MN599039 | MN599040 |
High quality DNA was sheared to yield fragments less than or equal to 800 bp, and fragment quality was checked using Agilent Bioanalyzer 2100 (Agilent Technologies). The 500 bp short-insert length paired-end library was prepared and sequenced on an Illumina HiSeq X10 to obtain reads of 150 bp at Beijing Genomics Institute (BGI, Wuhan, China).
Raw reads were first screened for Phred score < 30 to remove low quality sequences. All remaining reads were assembled into contigs implemented in the CLC Genomics Workbench (CLC Inc. Aarhus, Denmark). The parameters set in CLC were as follows: 200 bp for minimum contig length, 3 for deletion and insertion costs, bubble size of 98, 0.9 for length fraction and similarity fraction, 2 for mismatch cost. Principal contigs representing the plastome were then screened from the total contigs using a BLAST (NCBI BLAST v.2.2.31) search, with the published plastome of Celtis biondii (GenBank accession number: MH118119) as a reference. Mapped contigs were oriented and ordered according to the reference genome to yield the draft genomes, and plastomes were finalized by re-mapping cleaned reads to the draft genomes.
We used Geneious R11 (Kearse et al., 2012) to annotate the plastomes, and putative starts, stops, and intron positions were identified by comparison with homologous genes of the reference genome. The tRNA genes were verified with tRNAscan-SE v.1.21 (Schattner et al., 2005) using the default setting. We drew the graphical map of the annotated circular plastomes using the OrganellarGenomeDRAW program (OGDRAW, Lohse et al., 2013).
2.3. Comparative analysis of plastomes in CeltisTo analyze sequence variation within the genus, multiple sequence alignments of the seven Celtis plastomes were performed in MAFFT v.7.017 (Katoh and Standley, 2013) with standard parameters. Alignments were visually inspected and manually adjusted in Geneious R11. Sequence identities of the seven Celtis plastomes were plotted using the mVISTA program in LAGAN mode (Frazer et al., 2004). Plastid DNA rearrangement analyses were performed via whole genome alignment in Mauve (Darling et al., 2004).
2.4. Genomic resources development for CeltisTo identify plastid hotspot regions within the genus Celtis, multiple alignments of the seven plastomes were performed using MAFFT v.7.017. Nucleotide diversity (Pi) was determined by calculating the total number of mutations (Eta) and average number of nucleotide differences (K) using DnaSP v.5.0 (Librado and Rozas, 2009).
Plastid and mitochondrial sequences of the Celtis were removed from the assembled contigs using NCBI BLAST with the plastome of C. biondii and mitochondrial genome of Cannabis sativa L (GenBank accession number: KU310670) as references. Candidate polymorphic nSSRs within the genus Celtis were screened based on multiple assembled sequences using the CandiSSR v20170602 software (Xia et al., 2016) set at default parameters. Primers for each target SSR were automatically designed based on the Primer3 package built-in installation to the pipeline (Untergasser et al., 2012).
For targeted sequence capture, we used probes from 353 putative single-copy protein-coding genes from 42 angiosperms (Johnson et al., 2018), which have been shown to be universal in 283 species (the information on sequences are published at github. com/mossmatters/Angiosperms353). Our cleaned sequence data for each Celtis species was searched using pipeline HybPiper v.1.2 (Johnson et al., 2016) with default parameters. Under ideal conditions, searches identify only single contigs for each Celtis gene sequence.
2.5. Phylogenetic analysisPhylogenetic inferences were conducted using Bayesian inference (BI) and Maximum-Likelihood (ML) methods for three different data sets: (1) a combined matrix of four plastid regions (psbA-trnH, matK, rbcL and trnL-trnF; mostly downloaded from GenBank, see Table S1) for 32 Celtis species (to reduce missing data, we only used species with at least two regions); (2) whole plastome sequences of seven Celtis accessions; (3) 78 protein-coding gene sequences commonly shared among the seven accessions. Two species from Cannabaceae, Trema orientalis (L.) Blume and Pteroceltis tatarinowii Maxim., were used as outgroups according to Zhang et al. (2018).
The best-fit nucleotide substitution model (GTR + G for three datasets) was determined by jModelTest v.2.1.4 (Posada, 2008). ML analysis was performed with RAxML-HPC v.8.1.11 on the CIPRES cluster (Miller et al., 2010), with 1000 bootstrap replicates. BI analysis was performed using MrBayes v.3.2.3 (Ronquist and Huelsenbeck, 2003). The Markov chain Monte Carlo (MCMC) algorithm was run under the parameters set as follows: five million generations, every 1000 generations for tree sampling, and convergence was determined when the average standard deviation of split frequencies reached 0.01 or less. The first 25% of the trees were discarded as a burn-in to generate the consensus tree.
3. Results and discussion 3.1. Illumina sequencing and genome featuresMore than 10 Gbp of high-quality data with clean reads (Q30 > 95%) were generated from the Illumina sequencing platform for each species. Between 419, 052 (Celtis koraiensis) to 527, 009 (C. cerasifera) contigs were generated after de novo assembly. All plastomes were reconstructed from three initial contigs corresponding to the large single-copy region (LSC), small single-copy region (SSC), and inverted repeat regions (IRB/IRA), with no Ns or gaps detected. All five plastome sequences have been submitted to GenBank (accession numbers shown in Table 1).
Plastomes of all five Celtis species possessed a typical quadripartite structure similar to the majority of land plant plastid genomes (Li et al., 2017; Liu et al., 2017). The genome size ranged from 158, 989 bp in C. biondii to 159, 082 bp in C. koraiensis (Fig. 1, Table 1), consisting of two copies of IR regions (26, 861–26, 862 bp) separated by a LSC region (86, 075–86, 171 bp) and an SSC region (19, 129–19, 190 bp). All five plastomes contained 112 unique genes, including 78 protein-coding genes, 30 tRNA genes, and four rRNA genes with 20 genes duplicated in IR regions (Table 2). Of the 112 genes, nine protein-coding genes and six tRNA genes contained one intron, and three protein-coding genes (rps12, clpP and ycf3) contained two introns.

|
| Fig. 1 Plastome maps of five Celtis species. Genes inside the circle are transcribed clockwise, gene outside are transcribed counter-clockwise. The light gray inner circle corresponds to AT content, the dark gray to GC content. Genes belonging to different functional groups are shown in different colors; see the legend for groups. |
| Category | Group of gene | Name of gene | |||
| Self-replication | Ribosomal RNA genes | rrn4.5a | rrn5a | rrn16a | rrn23a |
| Transfer RNA genes | trnA-UGCa* | trnC-GCA | trnD-GUC | trnE-UUC | |
| trnF-GAA | trnfM-CAU | trnG-GCC | trnG-UCC* | ||
| trnH-GUG | trnI-CAUa | trnI-GAUa* | trnK-UUU* | ||
| trnL-CAAa | trnL-UAA* | trnL-UAG | trnM-CAU | ||
| trnN-GUUa | trnP-UGG | trnQ-UUG | trnR-ACGa | ||
| trnR-UCU | trnS-GCU | trnS-GGA | trnS-UGA | ||
| trnT-GGU | trnT-UGU | trnV-GACa | trnV-UAC* | ||
| trnW-CCA | trnY-GUA | ||||
| Small subunit of ribosome | rps2 | rps3 | rps4 | rps7a | |
| rps8 | rps11 | rps12a, b** | rps14 | ||
| rps15 | rps16* | rps18 | rps19a | ||
| Large subunit of ribosome | rpl2a* | rpl14 | rpl16* | rpl20 | |
| rpl22a | rpl23a | rpl32 | rpl33 | ||
| rpl36 | |||||
| RNA polymerase subunits | rpoA | rpoB | rpoC1* | rpoC2 | |
| Photosynthesis | Subunits of photosystem Ⅰ | psaA | psaB | psaC | psaI |
| psaJ | ycf3** | ||||
| Subunits of photosystem Ⅱ | psbA | psbB | psbC | psbD | |
| psbE | psbF | psbH | psbI | ||
| psbJ | psbK | psbL | psbM | ||
| psbN | psbT | psbZ | |||
| Subunits of cytochrome | petA | petB* | petD* | petG | |
| petL | petN | ||||
| Subunits of ATP synthase | atpA | atpB | atpE | atpF* | |
| atpH | atpI | ||||
| Large subunit of Rubisco | rbcL | ||||
| Subunits of NADH Dehydrogenase | ndhA* | ndhBa* | ndhC | ndhD | |
| ndhE | ndhF | ndhG | ndhH | ||
| ndhI | ndhJ | ndhK | |||
| Other genes | Maturase | matK | |||
| Envelope membrane protein | cemA | ||||
| Subunit of acetyl-CoA | accD | ||||
| C-type cytochrome synthesis gene | ccsA | ||||
| Protease | clpP** | ||||
| Unknown function | Conserved open reading frames | ycf1a (part) | ycf2a | ycf4 | |
|
*Indicates one intron containing genes.
**Indicates two intron containing genes. a Two gene copies in IRs. b Gene divided into two independent transcription units. |
|||||
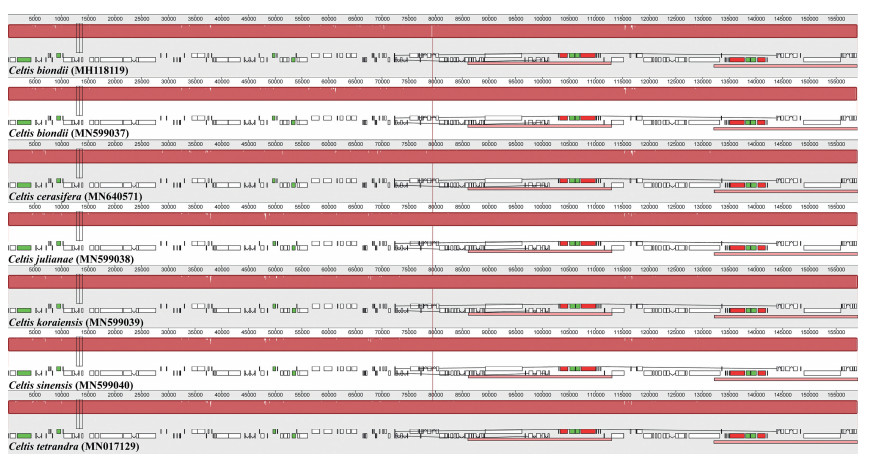
Multiple sequence alignment of seven Celtis plastomes showed no evidence of genome structure variation (e.g., gene loss, translocation, inversions, or genome rearrangement) (Fig. 2). Plastome sequences were relatively conserved especially the IR regions were more conserved than both single copy regions (Fig. 3).
|
| Fig. 2 Mauve alignment of seven Celtis plastomes. Within each of the alignments, local collinear blocks are represented by blocks of the same color connected by lines. |
|
| Fig. 3 Visualization of alignment of seven Celtis plastome sequences with C. biondii as a reference. The horizontal axis indicates the coordinates within the plastome. The vertical scale indicates the percentage of identity, ranging from 50 to 100%. Genome regions are color coded as protein coding, intron, mRNA, and conserved non-coding sequences (CNS). |
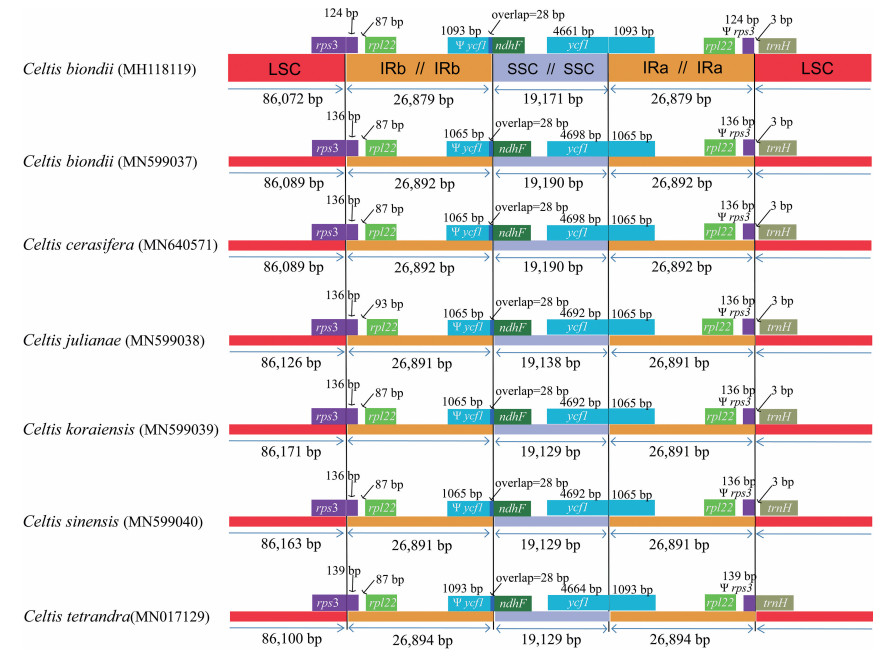
Most angiosperm plastomes range between 135 and 160 kb (Palmer, 1985). The most typical cause of variation in the length of plastomes is the contraction or expansion of the IR into or out of adjacent single copy regions (Plunkett and Downie, 1999). Therefore, we compared the exact IR border positions of seven Celtis plastomes and the positions of adjacent genes (Fig. 4). The genes rps3-rpl22-trnH and ycf1-ndhF were located in the boundaries of LSC/IR and SSC/IR regions. The ycf1 gene spanned the SSC/IRA region. In C. biondii (MH118119) and C. tetrandra, the pseudogene fragment of ψycf1 was 1093 bp, whereas in the remaining Celtis plastomes it was 1065 bp. In all seven Celtis plastomes, the ndhF gene shared a 28 bp sequence with the ψycf1 gene. The rps3 gene spanned the LSC/IRB region, resulting in 124 bp to 139 bp nucleotides located in IRB region. In C. julianae, the rpl22 gene was divided from the LSC/IRB boundary by a 93 bp spacer and this spacer was 87 bp in the remaining species. For all species except C. tetrandra, the trnH-GUG gene was separated from the IRA/LSC boundary by a spacer of 3 bp. Compared to plastomes of other genera within Cannabaceae (Zhang et al., 2018), the genome size and genome boundary structure of Celtis plastomes are relatively conserved.
|
| Fig. 4 Comparison of the borders of large single-copy (LSC), small single-copy (SSC), and inverted repeat (IR) regions among the seven Celtis chloroplast genomes. |
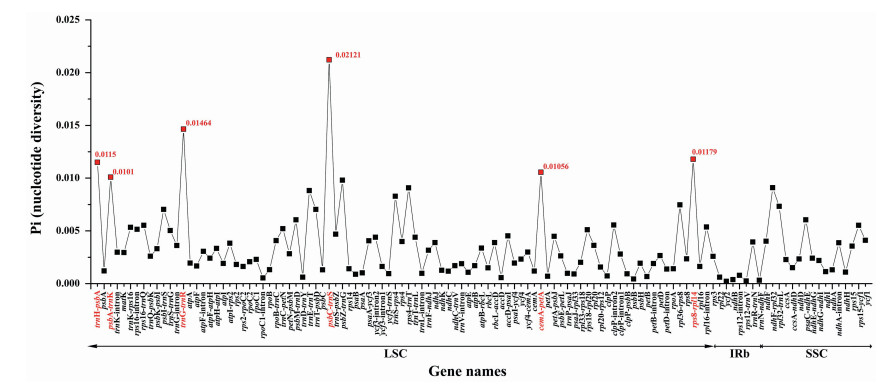
Although plastomes are usually an effective tool to investigate species divergence, identify species and trace demographic history (Thomson et al., 2010), traditional screening approaches are time consuming and inefficient (Alkan et al., 2011). Here, we screened divergence hotspots in the plastomes of seven Celtis accessions by comparing coding genes, non-coding regions, and intron regions. A total of 117 loci (56 inter-genic spacers, 45 coding genes, and 16 intron regions) with a length more than 200 bp were generated (Fig. 5). Nucleotide diversity (Pi) values for each locus ranged from 0.00022 (ycf2) to 0.02121 (psbC-trnS). Six of these loci with the Pi ≥ 0.01, including trnH-psbA, psbA-trnK, trnG-trnR, psbC-trnS, cemA-petA and rps8-rpl14, showed relative high nucleotide diversity values and can be used as highly informative phylogenetic markers for the genus Celtis or higher taxonomic levels.
|
| Fig. 5 Comparative analysis of the nucleotide variability (Pi) values within the genus Celtis. Genes surpassing the threshold (Pi ≥ 0.01) are highlighted in red. |
Genomic SSR (gSSR) markers have higher levels of polymorphism than EST-SSRs, and have thus garnered more attention (Bae et al., 2015). In this study, a total of 11, 559 candidate polymorphic nSSRs were identified for the genus Celtis. After discarding loci with either no available primers or sequence similarity < 99%, we obtained 4097 polymorphic nSSRs with a standard deviation between 0.4 and 3.7 (Table S2, Table S3). Of these polymorphic nSSRs, di- (2940), tri- (997), tetra- (140), penta- (13) and hexanucleotides (7) accounted for 71.76%, 24.33%, 3.42%, 0.32% and 0.17%, respectively (Fig. 6). Dinucleotide repeats included AT/TA (64.56%), CT/TC (15.56%) and AG/GA (15.07%) motifs. SSRs in genus Celtis are strongly biased towards AT-rich repeat motifs, not only in dinucleotides but also in tri-, tetra-, penta- and hexanucleotide repeats.

|
| Fig. 6 The distribution of polymorphic nucleotide simple sequence repeats (nSSRs) for the genus Celtis. (a), (b), (c), (d) and (e) represent di-, tri-, tetra-, penta- and hexanucleotide repeats, respectively. |
Several plastid markers are frequently used for phylogenetic studies, including single genes (Soltis et al., 1993; Cameron et al., 1999), whole plastid exomes (Gitzendanner et al., 2018; Medina et al., 2018) or whole plastome sequences (Bernhardt et al., 2017; Liu et al., 2017). Importantly, phylogenetic relationships based on plastome data alone may not reflect the evolutionary divergence of plants in Celtis, which have likely undergone extensive interspecific hybridization (Fernald, 1950). We used the HybPiper pipeline to identify putative single-copy genes for phylogenetic analysis of genus Celtis. In total, 353 target genes were successfully mapped from the raw clean reads, and contigs were formed for C. biondii (11), C. cerasifera (14), C. julianae (26), C. koraiensis (11) and C. sinensis (27) (Fig. 7; Table S4). We recovered coding sequences with a length of at least 50% of the target length for C. biondii (2), C. cerasifera (4), C. julianae (8), C. koraiensis (3) and C. sinensis (6). In addition, we recovered three coding sequences (TJLC, THHD and YKQR) in C. sinensis longer than the corresponding target genes (Table S4). Although the gene recovery rate appears relatively low and no recovered genes were shared in the five sequenced Celtis species, we were still able to obtain homologous genes within the genus through PCR amplification using universal primers. In combination with plastome data, these genes will provide useful molecular tools for further studies on the history of species divergence in Celtis.

|
| Fig. 7 Heatmap of gene recovery efficiency. Each row represents each Celtis species, and each column is one gene. Shading indicates the percentage of the target length recovered. |
ML and BI analyses based on a combined matrix of four plastid regions (3940 bp) for all sampled species generated identical trees (Fig. 8). The 32 Celtis species were divided into five major clades, but the ML bootstrap support (BS) and BI posterior probability (PP) for most clades were weak. Nine of the eleven species distributed in China were assigned to two clades (Clade A and Clade D), and the six species with available plastomes were assigned to one subclade of Clade A.

|
| Fig. 8 Phylogenetic tree reconstruction of 32 Celtis species using maximum likelihood (ML) based on a combined matrix of four plastid regions. The inset topology in the upper left shows the relative branch lengths in per-site substitutions. Numbers above the branches represent ML bootstrap/Bayesian posterior probability (BS/PP). Hyphens indicate a bootstrap value or posterior probability < 50%. |
We also used a plastome data set and a concatenated matrix of 78 shared protein-coding genes for ML and BI analyses. The length of aligned whole plastome sequences (nine individuals) was 166, 417 bp, and contained 9826 polymorphic sites, which can be further divided into 6548 singleton variable sites and 3278 parsimony informative sites. The concatenated matrix of 78 shared protein-coding genes was 68, 849 bp in length, comprising 2434 singleton variable sites and 1325 parsimony informative sites. The tree topologies recovered from both the whole plastome and the concatenated data sets were completely coincident (Fig. 9). The monophyly of Celtis had maximal support (MLBS = 100%, BIPP = 1). Within Celtis, C. biondii was first to diverge from the six representative species, and the remaining five species formed the topology ((((C. koraiensis, C. sinensis), C. tetrandra), C. julianae), C. cerasifera), with strong support.
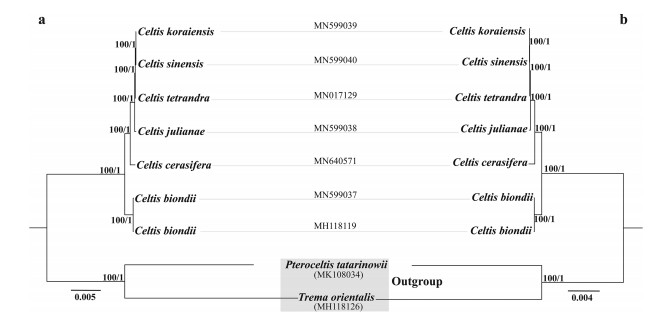
|
| Fig. 9 Phylogenetic tree reconstruction of seven Celtis accessions using maximum likelihood (ML) based on a) whole plastome sequences and b) 78 shared protein-coding genes. Numbers above the lines represent ML bootstrap values/BI posterior probability. |
To eliminate sampling interference, we removed all Celtis species except for six species (seven accessions), which were retained to reconstruct the phylogenetic tree based on four plastid regions (Fig. S1). The interspecific relationships of the six Celtis species resolved by plastomes and 78 shared protein-coding genes were consistent with the results recovered by the four combined plastid regions but with higher BS/PP support values.
In this study, we used plastome data to reconstruct the phylogenetic relationships of six Celtis species and compared these phylogenies to those generated using four common barcoding regions. The tree topologies inferred from the whole plastomes and 78 shared protein-coding genes are identical, and every branch of these trees is supported by full BS/PP values (Fig. 9). We also found that the phylogenetic relationships of the six species were consistent with that revealed by the combined matrix of four plastid regions (Fig. S1), although these data generated trees with much lower BS/PP values. Thus, our findings indicate that whole plastome or CDS genes have more informative sites and are more effective at resolving the phylogeny of Celtis.
4. ConclusionsIn this study, five Celtis species were sampled for genome skimming. Combined with two published plastomes, six Celtis species (seven accessions) were used to generate abundant genetic resources, including plastid hotspots, polymorphic nuclear SSRs and low or single copy gene fragments. All the sequenced plastomes had a typical quadripartite structure with similar size and organization to other sequenced angiosperms. In addition, phylogenetic relationships of Celtis species were well-resolved based on whole plastomes or commonly shared protein-coding genes. Our phylogenetic tree of the genus Celtis and the genetic resources developed herein will benefit further studies of phylogeny, population genetics, and conservation biology for this important woody genus.
Author contributionsPL and LXL designed the study. LXL conducted the sampling. LXL and YHZ produced and analyzed the data. LXL and PL wrote the manuscript. YHZ revised the manuscript. All authors approved the final manuscript.
Declaration of competing interestThe authors declare that they have no competing interests.
AcknowledgmentsWe sincerely thank Dr. James R.P. Worth and Ryan A. Folk for revising the manuscript. This research was supported by the National Natural Science Foundation of China (Grant Nos. 31900188, 31970225), Natural Science Foundation of Zhejiang Province (Grant No. LY19C030007).
Appendix A. Supplementary dataSupplementary data to this article can be found online at https://doi.org/10.1016/j.pld.2020.09.005.
Alkan C., Sajjadian S., Eichler E.E., 2011. Limitations of next-generation genome sequence assembly. Nat. Methods, 8: 61-65. DOI:10.1038/nmeth.1527 |
Angiosperm Phylogeny Group Ⅲ, 2009. An update of the angiosperm phylogeny group classification for the orders and families of flowering plants: APG Ⅲ. Bot.J. Linn. Soc, 161: 105-121. DOI:10.1111/j.1095-8339.2009.00996.x |
Bae K.M., Sim S.C., Hong J.H., et al, 2015. Development of genomic SSR markers and genetic diversity analysis in cultivated radish (Raphanus sativus L.). Hortic.Environ. Biote, 56: 216-224. DOI:10.1007/s13580-015-0089-y |
Baldauf S.L., Roger A., Wenk-Siefert I., et al, 2000. A kingdom-level phylogeny of eukaryotes based on combined protein data. Science, 290: 972-977. DOI:10.1126/science.290.5493.972 |
Bernhardt N., Brassac J., Kilian B., et al, 2017. Dated tribe-wide whole chloroplast genome phylogeny indicates recurrent hybridizations within Triticeae. BMC Evol. Biol, 17: 141. DOI:10.1186/s12862-017-0989-9 |
Besnard G., Christin P.A., Malé P.J.G., et al, 2013. Phylogenomics and taxonomy of Lecomtelleae (Poaceae), an isolated panicoid lineage from Madagascar. Ann.Bot, 112: 1057-1066. DOI:10.1093/aob/mct174 |
Cameron K.M., Chase M.W., Whitten W.M., et al, 1999. A phylogenetic analysis of the Orchidaceae: evidence from rbcL nucleotide sequences. Am. J. Bot, 86: 208-224. DOI:10.2307/2656938 |
Chen Z.L., Lu Y.F., Zhang W.B., et al, 2017. Celtis neglecta (Cannabaceae), a new species from Zhejiang, eastern China. Phytotaxa, 298: 55-64. DOI:10.11646/phytotaxa.298.1.5 |
Correll D.S., Johnston M.C., 1970. Manual of the vascular plants of Texas. Contributions From Texas Res. Foundation A Series Botanical Studies: 6. |
Cronn R., Knaus B.J., Liston A., et al, 2012. Targeted enrichment strategies for nextgeneration plant biology. Am. J. Bot, 99: 291-311. DOI:10.3732/ajb.1100356 |
Darling A.C., Mau B., Blattner F.R., et al, 2004. Mauve: multiple alignment of conserved genomic sequence with rearrangements. Genome Res, 14: 1394-1403. DOI:10.1101/gr.2289704 |
Duvall M.R., Fisher A.E., Columbus J.T., et al, 2016. Phylogenomics and plastome evolution of the chloridoid grasses (Chloridoideae: poaceae). Int. J. Plant Sci, 177: 235-246. DOI:10.1086/684526 |
Fernald M.L., 1950. Gray's Manual of Botany. 8th New York: American Book Company.
|
Frazer K.A., Pachter L., Poliakov A., et al, 2004. VISTA: computational tools for comparative genomics. Nucleic Acids Res, 32: W273-W279. DOI:10.1093/nar/gkh458 |
Gitzendanner M.A., Soltis P.S., Wong G.K.S., et al, 2018. Plastid phylogenomic analysis of green plants: a billion years of evolutionary history. Am. J. Bot, 105: 291-301. DOI:10.1002/ajb2.1048 |
Givnish T.J., Spalink D., Ames M., et al, 2015. Orchid phylogenomics and multiple drivers of their extraordinary diversification. P. Roy. Soc. B-Biol. Sci, 282: 20151553. |
Gruenstaeudl M., Santos-Guerra A., Jansen R.K., 2013. Phylogenetic analyses of Tolpis Adans. (Asteraceae) reveal patterns of adaptive radiation, multiple colonization and interspecific hybridization. Cladistics, 29: 416-434. DOI:10.1111/cla.12005 |
Gu C., Ma L., Wu Z., et al, 2019. Comparative analyses of chloroplast genomes from 22 Lythraceae species: inferences for phylogenetic relationships and genome evolution within Myrtales. BMC Plant Biol, 19: 281. DOI:10.1186/s12870-019-1870-3 |
Hooker, J.D., 1885. Flora of British India, vol. 7. L. Reeve & Co, London, Ashford, Kent.
|
Hwang B.Y., Chai H.B., Kardono L.B., et al, 2003. Cytotoxic triterpenes from the twigs of Celtis philippinensis. Phytochemistry, 62: 197-201. DOI:10.1016/S0031-9422(02)00520-4 |
Jansen R.K., Cai Z., Raubeson L.A., et al, 2007. Analysis of 81 genes from 64 plastid genomes resolves relationships in angiosperms and identifies genome-scale evolutionary patterns. P. Natl. Acad. Sci. U.S.A, 104: 19369-19374. DOI:10.1073/pnas.0709121104 |
Johnson M.A., Pillon Y., Sakishima T., et al, 2019. Multiple colonizations, hybridization and uneven diversification in Cyrtandra (Gesneriaceae) lineages on Hawai'i Island. J. Biogeogr, 46: 1178-1196. DOI:10.1111/jbi.13567 |
Johnson M.G., Gardner E.M., Liu Y., et al, 2016. HybPiper: extracting coding sequence and introns for phylogenetics from high-throughput sequencing reads using target enrichment. Appl. Plant Sci, 4: 1600016. DOI:10.3732/apps.1600016 |
Johnson M.G., Pokorny L., Dodsworth S., et al, 2018. A universal probe set for targeted sequencing of 353 nuclear genes from any flowering plant designed using k-medoids clustering. Syst. Biol, 68: 594-606. |
Kane N., Sveinsson S., Dempewolf H., et al, 2012. Ultra-barcoding in cacao (Theobroma spp.; Malvaceae) using whole chloroplast genomes and nuclear ribosomal DNA. Am. J. Bot, 99: 320-329. DOI:10.3732/ajb.1100570 |
Katoh K., Standley D.M., 2013. MAFFT multiple sequence alignment software version 7:improvements in performance and usability. Mol. Biol. Evol, 30: 772-780. DOI:10.1093/molbev/mst010 |
Kearse M., Moir R., Wilson A., et al, 2012. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics, 28: 1647-1649. DOI:10.1093/bioinformatics/bts199 |
Keerler, H.L., 1902. Our Native Trees and How to Identify Them; A Popular Study of Their Habits and Their Peculiarities. Scribner, New York.
|
Kim D.K., Lim J.P., Kim J.W., et al, 2005. Antitumor and antiinflammatory constituents from Celtis sinensis. Arch Pharm. Res. (Seoul), 28: 39-43. DOI:10.1007/BF02975133 |
Krajicek J.E., Williams R.D., 1990. Celtis occidentalis L. Hackberry. RM Bums and BH Honkala, tech. coord. Silvics of North America, 2: 262-265. |
Lee S.C., Chang C.F., Ho K.Y., 2011. Genetic diversity and molecular discrimination of the closely related Taiwanese Ulmaceae species Celtis sinensis Persoon and Celtis formosana Hayata based on ISSR and ITS markers. Afr. J. Agric. Res, 6: 4760-4768. |
Li P., Lu R.S., Xu W.Q., et al, 2017. Comparative genomics and phylogenomics of east asian tulips (amana, liliaceae). Front. Plant Sci, 8: 451. |
Librado P., Rozas J., 2009. DnaSP v5:a software for comprehensive analysis of DNA polymorphism data. Bioinformatics, 25: 1451-1452. DOI:10.1093/bioinformatics/btp187 |
Liu L.X., Du Y.X., Folk R.A., et al, 2020. Plastome evolution in Saxifragaceae and multiple plastid capture events involing Heuchera and Tiarella. Front. Plant Sci, 11: 361. DOI:10.3389/fpls.2020.00361 |
Liu L.X., Wang Y.W., He P.Z., et al, 2018. Chloroplast genome analyses and genomic resource development for epilithic sister genera Oresitrophe and Mukdenia (Saxifragaceae), using genome skimming data. BMC Genom, 19: 235. DOI:10.1186/s12864-018-4633-x |
Liu L.X., Li R., Worth J.R.P., et al, 2017. The complete chloroplast genome of Chinese bayberry (Morella rubra, Myricaceae): implications for understanding the evolution of Fagales. Front. Plant Sci, 8: 968. DOI:10.3389/fpls.2017.00968 |
Lohse M., Drechsel O., Kahlau S., et al, 2013. OrganellarGenomeDRAW-a suite of tools for generating physical maps of plastid and mitochondrial genomes and visualizing expression data sets. Nucleic Acids Res, 41: W575-W581. DOI:10.1093/nar/gkt289 |
Malé P.J., Bardon L., Besnard G., et al, 2015. Genome skimming by shotgun sequencing helps resolve the phylogeny of a pantropical tree family. Mol. Ecol.Resour, 14: 966-975. |
Martins J.L., Rodrigues O.R., de Sousa F.B., et al, 2015. Medicinal species with gastroprotective activity found in the Brazilian Cerrado. Fund. Clin. Pharmacol, 29: 238-251. DOI:10.1111/fcp.12113 |
Mcpherson H., Merwe M.V.D., Delaney S.K., et al, 2013. Capturing chloroplast variation for molecular ecology studies: a simple next generation sequencing approach applied to a rainforest tree. BMC Ecol, 13: 8. DOI:10.1186/1472-6785-13-8 |
Medina R., Johnson M., Liu Y., et al, 2018. Evolutionary dynamism in bryophytes: phylogenomic inferences confirm rapid radiation in the moss family Funariaceae. Mol. Phylogenet. Evol, 120: 240-247. DOI:10.1016/j.ympev.2017.12.002 |
Miller, M.A., Pfeiffer, W., Schwartz, T., 2010. Creating the CIPRES Science Gateway for Inference of Large Phylogenetic Trees. In: Gateway Computing Environments Workshop (GCE), 2010. IEEE, pp. 1-8.
|
Moncalvo J.M., Vilgalys R., Redhead S.A., et al, 2002. One hundred and seventeen clades of euagarics. Mol. Phylogenet. Evol, 23: 357-400. DOI:10.1016/S1055-7903(02)00027-1 |
Moore M.J., Soltis P.S., Bell C.D., et al, 2010. Phylogenetic analysis of 83 plastid genes further resolves the early diversification of eudicots. P. Natl. Acad. Sci, 107: 4623-4628. DOI:10.1073/pnas.0907801107 |
Palmer J.D., 1985. Comparative organization of chloroplast genomes. Annu. Rev.Genet, 19: 325-354. DOI:10.1146/annurev.ge.19.120185.001545 |
Plunkett G.M., Downie S.R., 1999. Major lineages within Apiaceae subfamily Apioideae: a comparison of chloroplast restriction site and DNA sequence data. Am. J. Bot, 86: 1014-1026. DOI:10.2307/2656619 |
Posada D., 2008. jModelTest: phylogenetic model averaging. Mol. Biol. Evol, 25: 1253-1256. DOI:10.1093/molbev/msn083 |
Ronquist F., Huelsenbeck J., 2003. MrBayes: Bayesian phylogenetic inference under mixed models. Bioinformatics, 19: 1572-1574. DOI:10.1093/bioinformatics/btg180 |
Sattarian, A., 2006. Contribution to the Biosystematics of Celtis L. (Celtidaceae) with Special Emphasis on the African Species. Ph. D. dissertation. Wageningen Universitieit, Wageningen, Netherlands.
|
Schattner P., Brooks A.N., Lowe T.M., 2005. The tRNAscan-SE, snoscan and snoGPS web servers for the detection of tRNAs and snoRNAs. Nucleic Acids Res, 33: W686-W689. DOI:10.1093/nar/gki366 |
Soltis D., Soltis P., Rieseberg L.H., 1993. Molecular data and the dynamic nature of polyploidy. Crit. Rev. Plant Sci, 12: 243-273. DOI:10.1080/07352689309701903 |
Straub S.C., Parks M., Weitemier K., et al, 2012. Navigating the tip of the genomic iceberg: next-generation sequencing for plant systematics. Am. J. Bot, 99: 349-364. DOI:10.3732/ajb.1100335 |
Thomson R.C., Wang I.J., Johnson J.R., 2010. Genome-enabled development of DNA markers for ecology, evolution and conservation. Mol. Ecol, 19: 2184-2195. DOI:10.1111/j.1365-294X.2010.04650.x |
Untergasser A., Cutcutache I., Koressaar T., et al, 2012. Primer 3-new capabilities and interfaces. Nucleic Acids Res, 40: e115. DOI:10.1093/nar/gks596 |
Wang Y., Yuan X.L., Zhang J.F., 2019. The complete chloroplast genome sequence of Celtis tetrandra. Mitochondrial DNA B, 4: 3463-3464. DOI:10.1080/23802359.2019.1674705 |
Whittemore A.T., 2005. Genetic structure, lack of introgression, and taxonomic status in the Celtis laevigata-C. reticulata complex (Cannabaceae). Syst. Bot, 30: 809-817. DOI:10.1600/036364405775097897 |
Whittemore A.T., Townsend A.M., 2007. Hybridization and self-compatibility in Celtis: AFLP analysis of controlled crosses. J. Am. Soc. Hortic. Sci, 132: 268-373. |
Williams A.V., Miller J.T., Small I., et al, 2016. Integration of complete chloroplast genome sequences with small amplicon datasets improves phylogenetic resolution in Acacia. Mol. Phylogenet. Evol, 96: 1-8. DOI:10.1016/j.ympev.2015.11.021 |
Xia E.H., Yao Q.Y., Zhang H.B., et al, 2016. CandiSSR: an efficient pipeline used for identifying candidate polymorphic SSRs based on multiple assembled sequences. Front. Plant Sci, 6: 1171. |
Xu W.Q., Losh J., Chen C., et al, 2018. Comparative genomics of figworts (scrophularia, scrophulariaceae), with implications for the evolution of scrophularia and lamiales. J. Syst. Evol, 57: 55-65. |
Yang M.Q., van Velzen R., Bakker F.T., et al, 2013. Molecular phylogenetics and character evolution of Cannabaceae. Taxon, 62: 473-485. DOI:10.12705/623.9 |
Yu X.Q., Yang D., Guo C., et al, 2018. Plant phylogenomics based on genomepartitioning strategies: progress and prospects. Plant Divers, 40: 158-164. DOI:10.1016/j.pld.2018.06.005 |
Zhang H., Jin J., Moore M.J., et al, 2018. Plastome characteristics of Cannabaceae. Plant Divers, 40: 127-137. DOI:10.1016/j.pld.2018.04.003 |



