b. School of Life Science, Yunnan University, Kunming, 650091, China;
c. School of Traditional Chinese Medicine, Guangdong Pharmaceutical University, Guangzhou, 510006, China
East Asia, harboring about 600 endemic genera (Manchester et al., 2009), exhibits a high level of floristic endemism in seed plants. The significant endemism in East Asia flora can be attributed to the persistence of a large number of relict taxa (paleo-endemism), as well as the emergence of substantial novel lineages (neo-endemism) (Manchester et al., 2009, Lu et al., 2018). Representatives of paleo-endemism include Ginkgo L., Metasequoia Hu & W. C. Cheng, Cyclocarya Iljinsk., Davidia Baill., Tetracentron Oliv.; an example of a neo-endemic genus is Rodgersia A. Gray, which originated in Northeast Asia during the Pliocene and extensively diversified in southwest China during the Pleistocene (Ma et al., 2017). The occurrence of many neo-endemic lineages in East Asia is most likely contributed to the establishment of Asian monsoon in the Neocene (Lu et al., 2018, Wen et al., 2014), as the climatic changes created significant habitat heterogeneities (Jacques et al., 2011, Santosh, 2011, Wan et al., 2007, Yao et al., 2011, Zhang et al., 2012), which may have triggered extensive lineage diversification (Axelrod et al., 1998, Wu et al., 2005). Therefore, the evolutionary histories of these endemic lineages can provide useful insights into the origin and evolution of floristic endemism and broaden our understanding of the geological history and climatic oscillations in East Asia. In recent years, several evolutionary studies on such endemic genera have been presented (e.g., Appelhans et al., 2016, Deng et al., 2016, Hu et al., 2017, Ma et al., 2017, Xie et al., 2018). However, these case studies only represent a small fraction of the real diversity of East Asian endemic genera, and the evolutionary history of most East Asian endemic genera remains poorly understood.
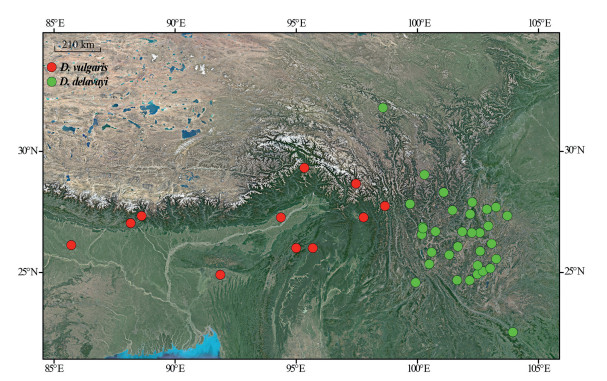
The genus Dobinea Buchanon-Hamilton ex D. Don is endemic to East Asia. The genus only includes two living species, namely, Dobinea delavayi Baillon and Dobinea vulgaris Buchanon-Hamilton ex D. Don (Fig. 1), which occur in southwest China and the Eastern Himalayas (Fig. 2) (Min and Barfod, 2008, Singh et al., 2011). D. delavayi is a perennial herb occurring in thickets or grasslands, whereas D. vulgaris is a shrub mainly growing in evergreen broad-leafed forests (Min and Barfod, 2008). Although their growth forms and habitats are different, they share distinctive morphological characters; specifically, the female flowers of both species lack petals and their single carpel is adnate to foliose bract. Because of this morphological distinctiveness, Dobinea is easy to recognize but difficult to classify taxonomically. Since the establishment of the genus, its taxonomic affinities have been controversial (Don, 1825, Engler, 1892, Morot, 1889, Pan et al., 2008, Radlkofer, 1888, Radlkofer, 1890). Based on the growth form and leaf morphology, Dobinea was firstly referred to the family Sapindaceae by Don (1825). Subsequently, Radlkofer (1888) transferred it to the family Anacardiaceae based on anatomical evidence. Due to their distinctive morphological characters (e.g., apetalous female flowers with a single carpel adnate to foliose bract), Morot (1889) established the family Podoonaceae to accommodate Dobinea. However, Radlkofer (1890) argued there were no solid evidence to maintain Podoonaceae as a separate family. Engler (1892) followed this suggestion and established the tribe Dobineeae within Anacardiaceae to accommodate this genus.

|
| Fig. 1 The morphological features of Dobinea. A, D. delavayi (Photo by Ting Zhang); B, D. vulgaris (Photo by Zhiling Dao). |
|
| Fig. 2 The species range of Dobinea delavayi and D. vulgaris. |
A high resolution and well-supported phylogeny could provide a valuable framework to resolve the disputes in the systematic placement of Dobinea. Based on plastid rbcL and nuclear Internal Transcribed Spacer (ITS) DNA sequences, the phylogenetic analyses by Pan et al. (2008) recovered Dobinea as a monophyletic unit within Anacardiaceae. Nevertheless, because both DNA regions lack sufficient sequence variation, the support and resolution of the phylogenetic trees are relatively low and the relationships between Dobinea and related taxa remain unresolved. Therefore, additional molecular data are needed to reconstruct a robust phylogeny.
Plastid genome (plastome) DNA sequences possess the highly variable characters required to reconstruct robust phylogenies in plants (Attigala et al., 2016, Kane et al., 2012, Tonti-Filippini et al., 2017). With the advent of the next-generation sequencing technologies, plastome DNA sequences have been widely used to resolve the historically difficult problems in plant phylogenetics (e.g., Huang et al., 2016, Jansen et al., 2007, Ji et al., 2019, Moore et al., 2007, Moore et al., 2010, Nock et al., 2011, Parks et al., 2009, Ruhsam et al., 2015, Yang et al., 2013, Yang et al., 2019). In this study, we firstly characterized the complete plastome DNA sequences for both extant species of Dobinea. Next, we investigated the phylogenetic relationships between Dobinea and related taxa based on the complete plastomes dataset. Finally, we dated the divergence of Dobinea to investigate the evolutionary history of the genus.
2. Material and methods 2.1. Plant material, DNA extraction, and plastome sequencingSamples of Dobinea delavayi and D. vulgaris were collected from the wild. The voucher information is shown in Supplementary Table S1. Genomic DNA was isolated from ~20 mg silica-gel-dried leaf tissues using the CTAB method (Doyle and Doyle, 1987). Genomic DNA was fragmented into 500 bp to construct a pair-end library. Illumina libraries were prepared according to manufacturer's protocol (Illumina, San Diego, CA, USA), and then sequenced on the Illumina HiSeq 2000 system.
2.2. Plastome assembly, annotation, and comparisonBoth reference-based and de novo assembly strategies were employed for plastome assembly. First, the Illumina raw data were used to assemble the complete plastome with GetOrganelle pipeline (Jin et al., 2018), using the plastome sequence of Mangifera indica L. (GenBank accession: NC_035239) as reference (Pan et al., 2008). Next, the plasotomes were assembled with the de novo assembler NOVOPlasty v3.8.3 (Dierckxsens et al., 2017) with a k-mer size of 31 and rbcL sequence from D. delavayi (EU123469) as the seed. To validate the accuracy of plastome assembly, the plastomes of a given species recovered by different assembly strategies were compared using Geneious v.10.2 (Kearse et al., 2012).
Initial annotations of plastomes were performed with the online software DOGMA (Wyman et al., 2004). The start and stop codons and intron positions were manually corrected according to the reference plastome sequence of M. indica in Geneious v.10.2 (Kearse et al., 2012). The tRNA genes were further verified by tRNAscan-SE 1.21 with the default parameters (Schattner et al., 2005). The annotated plastomes were illustrated using the online program OrganellarGenomeDRAW (Lohse et al., 2007). Sequence divergence between the plastomes of D. delavayi and D. vulgaris was compared using the mVISTA tool (Frazer et al., 2004), with the default parameters.
2.3. Phylogenetic analysesTo examine the phylogenetic position of Dobinea, we reconstructed a phylogenetic tree of the order Sapindales, using 55 complete plastome sequences publicly available, including representatives of two genera of Burseraceae, five genera of Meliaceae, two genera of Simaroubaceae, five genera of Anacardiaceae, ten genera of Sapindaceae, eight genera of Rutaceae, and the two newly sequenced Dobinea species (Table S2). Gossypium stocksii Mast. (Malvales) was used as the outgroup to root the phylogenetic tree, according to the results of Gadek et al. (1996). The alignment was performed with MAFFT software (Katoh et al., 2002), and adjusted manually when necessary. The ambiguous positions and insertions presented in one or few nucleotides in matrix were excluded from phylogenetic analyses. Unambiguous alignment was subjected to Maximum-Likelihood analyses (ML) and Bayesian Inference (BI). The ML phylogeny was reconstructed in the program RAxML-HPC BlackBox v.8.1.24 (Stamatakis, 2006) with the GTRGAMMAI model as the RAxML manual suggested. The bootstrap (BS) percentage for each branch was estimated by running 1000 bootstrap replicates. BI analysis was conducted with MrBayes v.3.2 (Ronquist and Huelsenbeck, 2003). The best-fit substitution model (GTR + I + G) for BI inference was determined by Modeltest v.3.7 (Posada and Buckley, 2004). Two independent Markov chains were run for 1, 000, 000 generations starting from a random tree and sampling every 100 generations. Trees for the first 25% were discarded as burn-in and the remainder were used to estimate the posterior probability values (PP). The run was finished when average standard deviation of split frequencies was below 0.01. Topologies of phylogenetic trees were visualized and edited in FigTree v.1.4.2 (Rambaut and Drummond, 2015).
2.4. Molecular datingWe estimated the divergence time of Dobinea with BEAST v.1.10.4 (Drummond and Rambaut, 2007). Two calibration points based on the fossil record and molecular dating for Anacardiaceae were used to constrain the phylogenetic tree. First, the divergence time of Anacardiaceae and Burseraceae was set to be at 70 ± 5.0 Ma (Bell et al., 2010, Estrada-Ruiz et al., 2010). Second, the node uniting Rhus and Pistacia was calibrated at 49.1 ± 2.1 Ma according to Yi et al. (2004). BEAST analysis was performed under the uncorrelated lognormal relaxed clock approach with a Yule tree prior and using GTR + I + G as the nucleotide substitution model. Markov Chain Monte Carlo chains were run for 10, 000, 000 generations with sampling every 1000 generations. The result was monitored using Tracer v.1.6 (Rambaut et al., 2014). Subsequently, a maximum credibility tree was generated in TreeAnnotator v.1.10.4 (Drummond et al., 2012) with the first 25% trees discarded as burn-in and visualized with FigTree v.1.4.2 (Rambaut and Drummond, 2015).
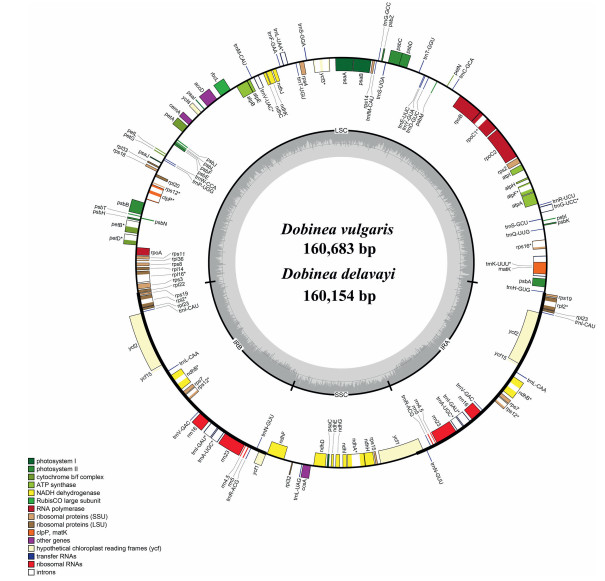
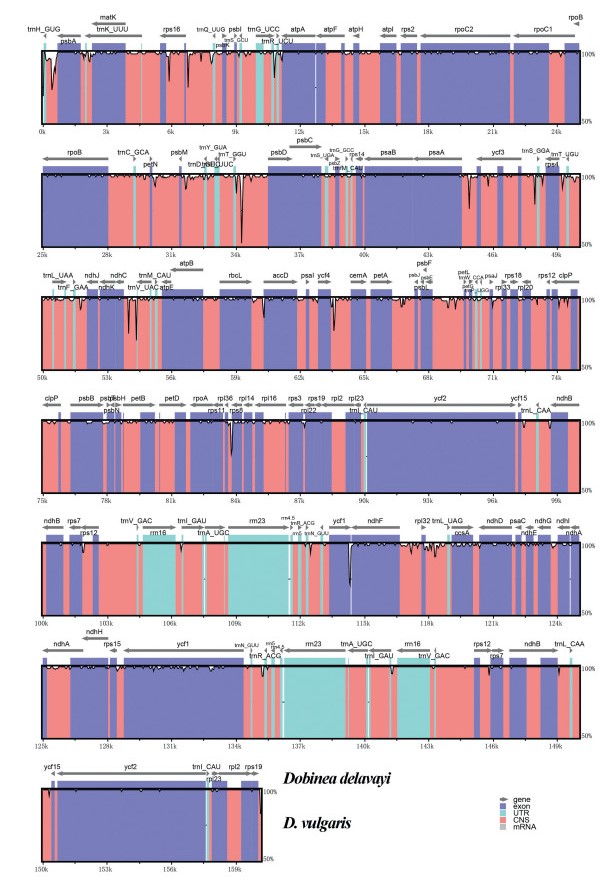
3. Results 3.1. Plastome features of DobineaIllumina sequencing obtained a total of 31, 505, 300 and 31, 529, 868 paired-end clean reads for Dobinea vulgaris and D. delavayi. Of those, 1, 475, 181 and 2, 028, 403 reads were mapped to the final assembly (Table S3). The overall sizes of assembled complete plastomes of D. vulgaris and D. delavayi were 160, 683 and 160, 154 bp. Both plastomes showed typically quadripartite structure, including a pair of inverted repeat regions (IRs, 26, 889 and 26, 759 bp) divided by the large single-copy region (LSC, 87, 962 and 87, 555 bp) and small single-copy region (SSC, 18, 943 and 19, 081 bp) (Fig. 3). The plastomes of Dobinea encoded 113 unique genes, including 79 protein-coding genes, 30 tRNAs, and 4 rRNA genes, of which twenty genes were duplicated in the IR regions. Among these unique genes, 12 protein-coding genes (atpF, ndhA, ndhB, petB, petD, rpl16, rpl2, rpoC1, rps12, rps16, clpP, and ycf3), and six tRNAs (trnA-UGC, trnG-UCC, trnI-GAU, trnK-UUU, trnL-UAA, and trnV-UAC) contained introns (Table S4). The whole plastome of D. vulgaris and D. delavayi exhibited 99.0% sequence identity. Plastome-wide comparative analysis using mVISTA identified only 568 variation sites among the 160, 961 alignment positions (Fig. 4).
|
| Fig. 3 Plastome map of the Dobinea. Genes presented in outside and inside of the circle are transcribed counterclockwise and clockwise, respectively. The colored bars indicate different gene functions. The dashed area in the inner circle denotes the GC content of the genome. LSC, large single-copy; SSC, small single-copy; IR, inverted repeat. |
|
| Fig. 4 Identity plot comparing the plastomes of two Dobinea species. Gray arrows indicate the position and direction of each gene. Black lines define the percentage of identity, ranging from 50 to 100%. Genome regions are color-coded as protein coding (exon), rRNA (UTR), tRNA (UTR), and conserved non-coding regions (CNS). |
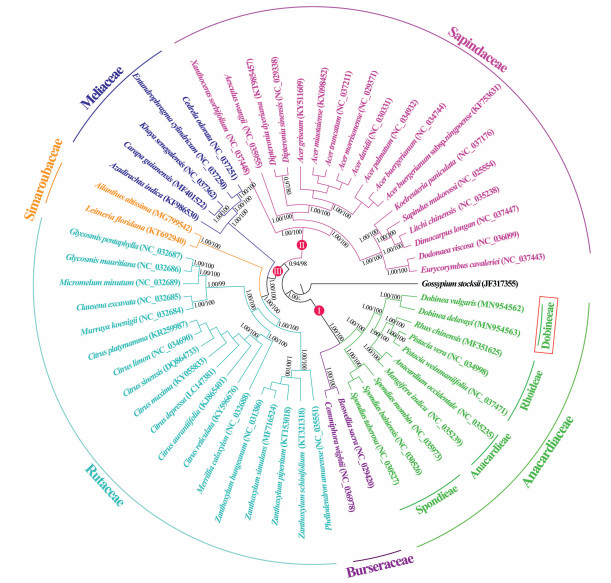
The tree topologies resulted from ML and BI analysis were identical. As indicated in Fig. 5, the fifty-five Sapindales taxa were fully resolved as three well-supported major clades. Clade Ⅰ contains Burseraceae and Anacardiaceae, which was placed at basal position of the phylogenetic trees (PP = 1.00, BS = 100). Clade Ⅱ is represented only by Sapindaceae (PP = 1.00, BS = 100). Clade Ⅲ included Meliaceae, Simaroubaceae and Rutaceae (PP = 1.00, BS = 100), in which the sister group Simaroubaceae and Rutaceae diverged from Meliaceae with strong support (PP = 1.00, BS = 100). In addition, all the Sapindales families (Sapindaceae, Meliaceae, Simaroubaceae, Rutaceae, Burseraceae, and Anacardiaceae) were recovered as fully supported (PP = 1.00, BS = 100) monophyletic lineages by our phylogenomic analyses.
|
| Fig. 5 Tree topology results from maximum likelihood (ML) and Bayesian inference (BI) analyses based on whole plastomes. Numbers represent Bayesian posterior probabilities (PP) and maximum likelihood bootstrap (BS). |
Within Anacardiaceae, four well-supported lineages (PP = 1.00, BS = 100), corresponding to the four tribes (Spondieae, Anacardieae, Rhoideae, and Dobineeae) recognized by Engler (1892), were recovered. Among them, the tribe Spondieae was the first clade that split from the rest of Anacardiaceae (PP = 1.00, BS = 100), and Dobinea (tribe Dobineeae) was sister to the clade including tribes Anacardieae and Rhoideae (PP = 1.00, BS = 100). Tree topology clearly indicates that the genus Dobinea was phylogenetically distinct from the members of Sapindaceae. Furthermore, molecular dating analysis revealed that D. delavayi and D. vulgaris diverged approximately 10.76 Ma (95% HPD: 5.10–17.76 Ma) (Fig. 6).
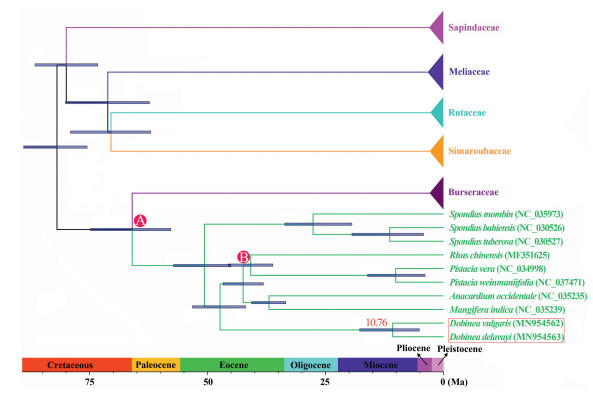
|
| Fig. 6 Divergence time estimation based on plastome DNA sequences. The red number above the tree branch indicates mean divergent age. Horizontal blue bars on each node indicate the 95% confidence interval of divergence time. Numbers on the Time Axis indicate million years ago (Ma). A: calibrated age = 70 ± 5.0 Ma; B: calibrated age = 49.1 ± 2.1 Ma. |
The order Sapindales, containing nine well-recognized families, i.e., Anacardiaceae, Biebersteiniaceae, Burseraceae, Kirkiaceae, Meliaceae, Nitrariaceae, Rutaceae, Sapindaceae and Simaroubaceae, is a member of core eudicots (Angiosperm phylogeny group, 2016). Although several studies have proposed inter-familial relationships within Sapindales (e.g., Clayton et al., 2007, Gadek et al., 1996, Lin et al., 2018, Muellner et al., 2003, Muellner et al., 2007, Muellner-Riehl et al., 2016), the relationships between the families Meliaceae, Rutaceae, Sapindaceae, and Simaroubaceae remain unresolved. The phylogenetic analyses of Muellner-Riehl et al. (2016) based on rbcL, atpB and trnL-trnF DNA sequences showed that Rutaceae was sister to the clade including Meliaceae and Simaroubaceae, with these together being sister to Sapindaceae. However, the phylogenetic analyses by Gadek et al. (1996) based on trnL-F sequence, as well as by Lin et al. (2018) based on 79 plastid genes revealed that Meliaceae was sister to the clade formed by Rutaceae and Simaroubaceae; Sapindaceae was not closely related to the clade consisting of Rutaceae, Meliaceae and Simaroubaceae. Our plastid phylogenomic analyses indicate that Meliaceae and the clade consisting of Simaroubaceae and Rutaceae robustly clustered into a group (PP = 1.00, BS = 100), which is, in turn, sister to Sapindaceae (PP = 0.94, BS = 98). The relationships recovered in this study are different from those of previous studies (Gadek et al., 1996, Lin et al., 2018, Muellner et al., 2007, Muellner-Riehl et al., 2016). With more variable characters and/or much larger taxon sample, our analyses provide new insights into the inter-familial relationships within Sapindales.
4.2. Phylogenetic position of DobineaSimilar to the previous study of Pan et al. (2008), the monophyly of Dobinea was strongly supported by the plastome-based phylogeny. In our analyses the genus was not phylogenetically related to members of the family Sapindaceae, but fully nested within Anacardiaceae with high support values (PP = 1.00, BS = 100). This robustly supports the taxonomic treatment of Dobinea as a member of Anacardiaceae (Radlkofer, 1888) rather than placing it into Sapindaceae (Don, 1825) or the separate family Podoonaceae (Forman, 1973, Hutchinson, 1973, Morot, 1889). The close relationships between Dobinea and other Anacardiaceae genera can be justified by the anatomical evidence: Dobinea harbor resin canals in the phloem, which is likely one of the morphological synapomorphies to recognize the family Anacardiaceae (Radlkofer, 1888). Nevertheless, Dobinea is morphologically distinctive in that female flowers lack petals and its single carpel is adnate to foliose bract, which is distinctly different from any other taxa within Anacardiaceae. This justifies the establishment of the tribe Dobineeae to accommodate the genus by Engler (1892).
The plastome-based phylogeny well resolved the inter-tribal relationships within Anacardiaceae. The tribes Spondieae, Anacardieae, and Rhoideae recognized by Engler (1892) were recovered as well-supported monophyletic units. The tribe Spondieae was recovered as the earliest diverged clade. This finding is congruent with previous studies that used rbcL and ITS sequences (Pan et al., 2008), two nuclear and seven plastid DNA regions (Xie et al., 2014), and two nuclear sequences and two plastid DNA regions (Yang et al., 2016). The basally branching position of Spondieae inferred in this study can be further supported by fossil evidence: the earliest fossil record for Anacardiaceae belongs to Spondieae, which was known from the Lower Eocene London Clay flora of southern England (Manchester et al., 2009). In addition, Dobineeae was sister to the clade including Anacardieae and Rhoideae. The affinity between them were also supported by the anatomical characters: Dobineeae and Anacardieae share similar carpel morphology and structure (Wannan and Quinn, 1991), while Dobineeae and Rhoideae have the same endocarp structure (Wannan and Quinn, 1990).
4.3. Species divergence within DobineaThe intensification of the Asian summer monsoon since the late Miocene established a humid climate in subtropical East Asia (Jacques et al., 2011, Wan et al., 2007, Zhang et al., 2012) and caused the expansion of forests in East Asia (Santosh, 2011, Yao et al., 2011); these processes played an essential role in driving and promoting a remarkable evolutionary radiation of plants (Chen et al., 2018, Lu et al., 2018, Sun and Wang, 2005, Wen et al., 2014). For instance, the intensification of Asian summer monsoon may have created largely unoccupied and favorable habitats for the genus Saussurea DC. (Asteraceae), which facilitated intensive lineage diversification in this genus (Wang et al., 2009). Previous studies also suggested that species diversification in Lepisorus (J. Sm.) Ching (Polypodiaceae) (Wang et al., 2012), Rheum L. (Polygonaceae) (Sun et al., 2012), Paris L. (Melanthiaceae) (Ji et al., 2019), Stewartia L. (Theaceae) (Lin et al., 2019) and Prinsepia Royle (Rosaceae) (Ma et al., 2019) was also correlated with the intensification of the monsoonal climate in East Asia.
Our molecular dating inferred that Dobinea species diverged 10.76 Ma (95% HPD: 5.10–17.76 Ma), around the late Miocene, when the Asian summer monsoon greatly intensified (An et al., 2001, Sun and Wang, 2005). This implies that the divergence between the two extant Dobinea species was most likely driven by the intensification of the Asian summer monsoon. The intensification of the Asian summer monsoon established distinct monsoon regimes in East Asia (Li et al., 2008, Yao et al., 2011). Since then, the Himalayan region (distribution area of D. vulgaris, Fig. 2) have been mainly governed by the Indian monsoon, whereas the Yunnan-Guizhou plateau (distribution area of D. delavayi, Fig. 2) have been affected by both the Indian and Pacific monsoons (Li et al., 2008, Yao et al., 2011). The divergence of monsoonal regimes may have triggered the vicariance between D. delavayi and D. vulgaris.
Author contributionsYJ conceived and designed the research. CL collected and analyzed the data. CL and YJ wrote the manuscript. YJ, LJ, SW and ZY discussed the results and revised the manuscript. All of the authors read and approved the manuscript.
Declaration of Competing InterestThe authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
AcknowledgementsThis study was financially supported by the Major program of National Natural Science Foundation of China (31590823). We are grateful to Drs. Zhiling Dao and Ting Zhang for providing photos of Dobinea species.
Appendix A. Supplementary dataSupplementary data to this article can be found online at http://tree.bio.ed.ac.uk/software/figtree/.
An Z.S., Kutzbach J.E., Prell W.L., Porter S.C., 2001. Evolution of Asian monsoons and phased uplift of the HimalayaeTibetan plateau since Late Miocene times. Nature, 411: 62-66. DOI:10.1038/35075035 |
Angiosperm phylogeny group, 2016. An update of the Angiosperm Phylogeny Group classification for the orders and families of flowering plants: APG Ⅳ. Bot. J. Linn.Soc, 181: 1-20. DOI:10.1111/boj.12385 |
Appelhans M.S., Krohm S., Manafzadeh S., Wen J., 2016. Phylogenetic placement of Psilopeganum, a rare monotypic genus of Rutaceae (the citrus family) endemic to China. J. Syst. Evol, 54(5): 535-544. DOI:10.1111/jse.12208 |
Attigala L., Wysocki W.P., Duvall M.R., Clark L.G., 2016. Phylogetic estimation and morphorlogical evolution of Arundinarieae (Bambusoideae: poaceae) based on plastome phylogenomic analysis. Mol. Phylogenet. Evol, 101: 111-121. DOI:10.1016/j.ympev.2016.05.008 |
Axelrod, D.I., Al-Shehbaz, I., Raven, P.H., 1998. History of the modern flora of China. In: Zhang, A.L., Wu, S.G. (Eds. ), Floristic Characteristics and Diversity of East Asian Plants. China Higher Education Press, Beijing, pp. 43-55.
|
Bell C.D., Soltis D.E., Soltis P.S., 2010. The age and diversification of the angiosperms re-revisited. Am. J. Bot, 97: 1296-1303. DOI:10.3732/ajb.0900346 |
Chen Y.S., Deng T., Zhou Z., Sun H., 2018. Is the East Asian flora ancient or not?. Natl. Sci. Rev, 5: 920-932. DOI:10.1093/nsr/nwx156 |
Clayton J.W., Fernando E.S., Soltis P.S., Soltis D.E., 2007. Molecular phylogeny of the tree-of-heaven family (Simaroubaceae) based on chloroplast and nuclear markers. Int. J. Plant Sci, 168(9): 1325-1339. DOI:10.1086/521796 |
Deng Y., Gao C., Xia N., Peng H., 2016. Wuacanthus (Acanthaceae), a new Chinese endemic genus segregated from Justicia (Acanthaceae). Plant Divers, 38(6): 312-321. DOI:10.1016/j.pld.2016.11.010 |
Dierckxsens N., Mardulyn P., Smits G., 2017. NOVOPlasty: de novo assembly of organelle genomes from whole genome data. Nucleic Acids Res, 45: e18. DOI:10.1093/nar/gkw1060 |
Don D., 1825. Prodromus florae Nepalensis etc. J. Gale. Londini: 249. |
Doyle J.J., Doyle J.L., 1987. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem. Bull, 19: 11-15. |
Drummond A.J., Rambaut A., 2007. BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evol. Biol, 7: 214. DOI:10.1186/1471-2148-7-214 |
Drummond A.J., Suchard M.A., Xie D., Rambaut A., 2012. Bayesian phylogenetics with BEAUti and the BEAST 1. 7. Mol. Biol. Evol, 29: 1969-1973. DOI:10.1093/molbev/mss075 |
Engler, A., 1892. Anacardiaceae. In: Engler, A., Prantl, K. (Eds. ), Die Naturlichen Pflanzenfamilien, vol. 3. Engelmann, Leipzig, pp. 137-178, 5.
|
Estrada-Ruiz E., Martinez-Cabrera H.I., Cevallos-Ferriz S.R.S., 2010. Upper cretaceous woods from the olmos formation (late campanian-early maastrichtian), coahuila, Mexico. Am. J. Bot, 97: 1179-1194. DOI:10.3732/ajb.0900234 |
Forman, L.L., 1973. Podoaceae. In: Airy Shaw, H.K. (Ed. ), A Dictionary of the Flowering Plants and Ferns, eighth ed. Cambridge University Press, Cambridge, p. 923.
|
Frazer K.A., Pachter L., Poliakov A., et al, 2004. VISTA: computational tools for comparative genomics. Nucleic Acids Res, 32: W273-W279. DOI:10.1093/nar/gkh458 |
Gadek P.A., Fernando E.S., Quinn C.J., et al, 1996. Sapindales: molecular delimitation and infraordinal groups. Am. J. Bot, 83(6): 802-811. DOI:10.1002/j.1537-2197.1996.tb12769.x |
Hu Y., Yan J., Feng X., et al, 2017. Characterization of the complete chloroplast genome of wheel wingnut (Cyclocarya paliurus), an endemic in China. Conserv.Genet. Resour, 9(2): 273-275. DOI:10.1007/s12686-016-0671-3 |
Huang Y., Li X., Yang Z., et al, 2016. Analysis of complete chloroplast genome sequences improves phylogenetic resolution in Paris (Melanthiaceae). Front.Plant Sci, 7: 1797. |
Hutchinson J., 1973. The Families of Flowering Plants. Oxford: Clarendon Press.
|
Jacques F.M.B., Guo S.X., Su T., et al, 2011. Quantitative reconstruction of the Late Miocene monsoon climates of southwest China: a case study of the Lincang flora from Yunnan Province. Palaeogeogr. Palaeoclimatol. Palaeoecol, 304: 318-327. DOI:10.1016/j.palaeo.2010.04.014 |
Jansen R.K., Cai Z., Raubeson L.A., et al, 2007. Analysis of 81 genes from 64 chloroplast genomes resolves relationships in angiosperms and identifies genome-scale evolutionary patterns. Proc. Natl. Acad. Sci. U.S.A, 104: 19369-19374. DOI:10.1073/pnas.0709121104 |
Ji Y., Yang L., Chase M.W., et al, 2019. Plastome phylogenomics, biogeography, and clade diversification of Paris (Melanthiaceae). BMC Plant Biol, 19(1): 1-14. DOI:10.1186/s12870-018-1600-2 |
Jin, J.J., Yu, W.B., Yang, J.B., et al., 2018. GetOrganelle: a simple and fast pipeline for de novo assembly of a complete circular chloroplast genome using genome skimming data bioRxiv 256479.
|
Kane N., Sveinsson S., Dempewolf H., et al, 2012. Ultra-barcoding in cacao (Theobroma spp.; Malvaceae) using whole chloroplast genomes and nuclear ribosomal DNA. Am. J. Bot, 99: 320-329. DOI:10.3732/ajb.1100570 |
Katoh K., Misawa K., Kuma K.I., Miyata T., 2002. MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res, 30: 3059-3066. DOI:10.1093/nar/gkf436 |
Kearse M., Moir R., Wilson A., et al, 2012. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics, 28: 1647-1649. DOI:10.1093/bioinformatics/bts199 |
Li F.J., Rousseau D.D., Wu N.Q., et al, 2008. Late Neogene evolution of the East Asian monsoon revealed by terrestrial mollusk record in western Chinese loess plateau: from winter to summer dominated subregime. Earth Planet Sci. Lett, 274: 439-447. DOI:10.1016/j.epsl.2008.07.038 |
Lin H.Y., Hao Y.J., Li J.H., et al, 2019. Phylogenomic conflict resulting from ancient introgression following species diversification in Stewartia s. l. (Theaceae). Mol.Phylogenet. Evol, 135: 1-11. DOI:10.1016/j.ympev.2019.02.018 |
Lin N., Moore M.J., Deng T., et al, 2018. Complete plastome sequencing from Toona(Meliaceae) and phylogenomic analyses within Sapindales. Appl. Plant Sci, 6(4): e1040. DOI:10.1002/aps3.1040 |
Lohse M., Drechsel O., Bock R., 2007. OrganellarGenomeDRAW (OGDRAW): a tool for the easy generation of high-quality custom graphical maps of plastid and mitochondrial genomes. Curr. Genet, 52: 267-274. DOI:10.1007/s00294-007-0161-y |
Lu L.M., Mao L.F., Yang T., et al, 2018. Evolutionary history of the angiosperm flora of China. Nature, 554(7691): 234-238. DOI:10.1038/nature25485 |
Ma X., Wang Z., Tian B., Sun H., 2019. Phylogeographic analyses of the East Asian endemic genus Prinsepia and the role of the East Asian monsoon system in shaping a north-south divergence pattern in China. Front. Genet, 10: 128. DOI:10.3389/fgene.2019.00128 |
Ma X.G., Sun W.G., Zhu W.D., Sun H., 2017. Resolving the phylogenetic relationships and evolutionary history of the East Asian endemic genus Rodgersia(Saxifragaceae) using multilocus data. Perspect. Plant Ecol, 25: 20-28. DOI:10.1016/j.ppees.2016.12.005 |
Manchester S.R., Chen Z.D., Lu A.M., Uemura K., 2009. Eastern Asian endemic seed plant genera and their paleogeographic history throughout the northern hemisphere. J. Syst. Evol, 47(1): 1-42. DOI:10.1111/j.1759-6831.2009.00001.x |
Min, T.L., Barfod, A., 2008. Dobinea. In: Wu, Z.Y., Raven, P.H. (Eds. ), Flora of China, vol. 11. Science Press and Missouri Botanical Garden Press, Beijing and St. Louis, p. 357.
|
Moore M.J., Bell C.D., Soltis P.S., Soltis D.E., 2007. Using plastid genome-scale data to resolve enigmatic relationships among basal angiosperms. Proc. Natl. Acad. Sci. Unit. States Am, 104: 19363-19368. DOI:10.1073/pnas.0708072104 |
Moore M.J., Soltis P.S., Bell C.D., et al, 2010. Phylogenetic analysis of 83 plastid genes further resolves the early diversification of eudicots. Proc. Natl. Acad. Sci.U.S.A, 107: 4623-4628. DOI:10.1073/pnas.0907801107 |
Morot L., 1889. Sur les affinités anatomique du genre Podoon. J. Bot. Paris, 3: 388-390. |
Muellner A.N., Samuel R., Johnson S.A., et al, 2003. Molecular phylogenetics of Meliaceae (Sapindales) based on nuclear and plastid DNA sequences. Am. J. Bot, 90(3): 471-480. DOI:10.3732/ajb.90.3.471 |
Muellner A.N., Vassiliades D.D., Renner S.S., 2007. Placing Biebersteiniaceae, a herbaceous clade of Sapindales, in a temporal and geographic context. Plant Systemat. Evol, 266(3-4): 233-252. DOI:10.1007/s00606-007-0546-x |
Muellner-Riehl A.N., Weeks A., Clayton J.W., et al, 2016. Molecular phylogenetics and molecular clock dating of Sapindales based on plastid rbcL, atpB and trnL-trnF DNA sequences. Taxon, 65(5): 1019-1036. DOI:10.12705/655.5 |
Nock C.J., Waters D.L.E., Edwards M.A., et al, 2011. Chloroplast genome sequences from total DNA for plant identification. Plant Biotechnol. J, 9: 328-333. DOI:10.1111/j.1467-7652.2010.00558.x |
Pan Y.Z., Gong X., Yang Y., 2008. Phylogenetic position of the genus Dobinea: evidence from nucleotide sequences of the chloroplast gene rbcL and the nuclear ribosomal ITS region. J. Syst. Evol, 46(4): 586-594. |
Parks M., Cronn R., Liston A., 2009. Increasing phylogenetic resolution at low taxonomic levels using massively parallel sequencing of chloroplast genomes. BMC Biol, 7: 1. DOI:10.1186/1741-7007-7-1 |
Posada D., Buckley T.R., 2004. Model selection and model averaging in phylogenetics: advantages of Akaike information criterion and Bayesian approaches over likelihood ratio tests. Syst. Biol, 53: 793-808. DOI:10.1080/10635150490522304 |
Radlkofer L.A., 1888. Ueber die Versetzung der Gattung Dobinea von den Acerineen zu den Anacardiaceen. S. B. Bayer. Akad. Wiss, 18: 385-395. |
Radlkofer L.A., 1890. Ueber die Gliederung der Familie der Sapindaceen. Sitz. Ber.Akad. Muenchen, 20: 105-319. |
Rambaut, A., Drummond, A., 2015. FigTree, version 1.4.2. Available from: http://tree.bio.ed.ac.uk/software/tracer/. (Accessed 6 November 2019).
|
Rambaut, A., Drummond, A., Suchard, M., 2014. Tracer, version 1.6. Available from: _aaaaaa_paichu__. (Accessed 6 November 2019).
|
Ronquist F., Huelsenbeck J.P., 2003. MrBayes 3:Bayesian phylogenetic inference under mixed models. Bioinformatics, 19: 1572-1574. DOI:10.1093/bioinformatics/btg180 |
Ruhsam M., Rai H.S., Mathews S., et al, 2015. Does complete plastid genome sequencing improve species discrimination and phylogenetic resolution in Araucaria? Mol. Ecol. Resour, 15(5): 1067-1078. DOI:10.1111/1755-0998.12375 |
Santosh M., 2011. History of supercontinents and its relation to the origin of Japanese islands. J. Geodyn, 120: 100-114. |
Schattner P., Brooks A.N., Lowe T.M., 2005. The tRNAscan-SE, snoscan and snoGPS web servers for the detection of tRNAs and snoRNAs. Nucleic Acids Res, 33: W686-W689. DOI:10.1093/nar/gki366 |
Singh V.N., Sinha B.K., Phukan S., Singh B., 2011. Dobinea Buchanon-Hamilton ex D. Don-a new generic record for Meghalaya and Nagaland, India. Pleione, 5(1): 198-200. |
Stamatakis A., 2006. RAxML-Ⅵ-HPC: maximum likelihood-based phylogenetic analysis with thousands of taxa and mixed models. Bioinformatics, 22: 2688-2690. DOI:10.1093/bioinformatics/btl446 |
Sun X.J., Wang P.X., 2005. How old is the Asian monsoon system? Palaeobotanical records from China. Palaeogeogr. Palaeoclimatol. Palaeoecol, 222: 181-222. DOI:10.1016/j.palaeo.2005.03.005 |
Sun Y., Wang A., Wan D., et al, 2012. Rapid radiation of Rheum (Polygonaceae) and parallel evolution of morphological traits. Mol. Phylogenet. Evol, 63(1): 150-158. DOI:10.1016/j.ympev.2012.01.002 |
Tonti-Filippini J., Nevill P.G., Dixon K., Small I., 2017. What can we do with 1000 plastid genomes?. Plant J, 90: 808-818. DOI:10.1111/tpj.13491 |
Wan S.M., Li A.C., Clift P.D., Stut J.B.W., 2007. Development of the East asian monsoon: mineralogical and sedimentologic records in the northern south China sea since 20 Ma. Palaeogeogr. Palaeoclimatol. Palaeoecol, 254: 561-582. DOI:10.1016/j.palaeo.2007.07.009 |
Wang L., Schneider H., Zhang X.C., Xiang Q.P., 2012. The rise of the Himalaya enforced the diversification of SE Asian ferns by altering the monsoon regimes. BMC Plant Biol, 12(1): 210. DOI:10.1186/1471-2229-12-210 |
Wang Y.J., Susanna A., Von Raab-Straube E., et al, 2009. Island-like radiation of Saussurea (asteraceae: cardueae) triggered by uplifts of the qinghaieTibetan plateau. Biol. J. Linn. Soc, 97(4): 893-903. DOI:10.1111/j.1095-8312.2009.01225.x |
Wannan B.S., Quinn C., 1990. Pericarp structure and generic affinities in the Anacardiaceae. Bot. J. Linn. Soc, 102: 225-252. DOI:10.1111/j.1095-8339.1990.tb01878.x |
Wannan B.S., Quinn C., 1991. Floral structure and evolution in the Anacardiaceae. Bot. J. Linn. Soc, 107: 349-385. DOI:10.1111/j.1095-8339.1991.tb00228.x |
Wen J., Zhang J.Q., Nie Z.L., et al, 2014. Evolutionary diversifications of plants on the qinghai-Tibetan plateau. Front. Genet, 5: 4. |
Wu Z.Y., Sun H., Zhou Z.K., Peng H., Li D.Z., 2005. Origin and differentiation of endemism in the flora of China. Acta Bot. Yunnanica, 27(6): 577-604. DOI:10.1007/s11515-007-0020-8 |
Wyman S.K., Jansen R.K., Boore J.L., 2004. Automatic annotation of organellar genomes with DOGMA. Bioinformatics, 20: 3252-3255. DOI:10.1093/bioinformatics/bth352 |
Xie D.F., Yu Y., Deng Y.Q., et al, 2018. Comparative analysis of the chloroplast genomes of the Chinese endemic genus Urophysa and their contribution to chloroplast phylogeny and adaptive evolution. Int. J. Mol. Sci, 19(7): 1847. DOI:10.3390/ijms19071847 |
Xie L., Yang Z.Y., Wen J., et al, 2014. Biogeographic history of Pistacia (Anacardiaceae), emphasizing the evolution of the Madrean-Tethyan and the eastern Asian-Tethyan disjunctions. Mol. Phylogenet. Evol, 77: 136-146. DOI:10.1016/j.ympev.2014.04.006 |
Yang J.B., Tang M., Li H.T., et al, 2013. Complete chloroplast genome of the genus Cymbidium: lights into the species identification, phylogenetic implications and population genetic analyses. BMC Evol. Biol, 13: 1. DOI:10.1186/1471-2148-13-1 |
Yang L., Yang Z., Liu C., et al, 2019. Chloroplast phylogenomic analysis provides insights into the evolution of the largest eukaryotic genome holder, Paris japonica (Melanthiaceae). BMC Plant Biol, 19(1): 293. DOI:10.1186/s12870-019-1879-7 |
Yang Y.Y., Meng Y., Wen J., et al, 2016. Phylogenetic analyses of Searsia (Anacardiaceae) from eastern Asia and its biogeographic disjunction with its African relatives. South Afr. J. Bot, 106: 129-136. DOI:10.1016/j.sajb.2016.05.021 |
Yao Y.F., Bruch A.A., Mosbrugger V., Li C.S., 2011. Quantitative reconstruction of Miocene climate patterns and evolution in southern China based on plant fossils. Palaeogeogr. Palaeoclimatol. Palaeoecol, 304: 291-307. DOI:10.1016/j.palaeo.2010.04.012 |
Yi T., Miller A.J., Wen J., 2004. Phylogenetic and biogeographic diversification of Rhus (Anacardiaceae) in the northern hemisphere. Mol. Phylogenet. Evol, 33(3): 861-879. DOI:10.1016/j.ympev.2004.07.006 |
Zhang Q.Q., Ferguson D.K., Mosbrugger V., et al, 2012. Vegetation and climatic changes of SW China in response to the uplift of Tibetan Plateau. Palaeogeogr.Palaeoclimatol. Palaeoecol, 363: 23-36. |



