b. College of Life Science, Yunnan University, Kunming, Yunnan, 650201, China;
c. Kunming College of Life Science, University of Chinese Academy of Sciences, Kunming, Yunnan, 650201, China
Plastome of most photosynthetic angiosperms consist of a pair of inverted repeat (IRs; usually around 25-kb) regions, a large single copy region (LSC; usually around 80-kb), and a small single copy region (SSC; usually around 20-kb) (Jansen, 2012). However, some plastomes show variable size due to the expansion and contraction of IR, and variable structure due to the inversion of genes or genome segments, the loss of IRs, the duplication and loss of the gene and genome segments, and the reduction of introns and genes (Wicke et al., 2011; Jansen, 2012). The GC content of the plastome, which is highly conserved, ranges between 34% and 40% (Jansen, 2012).
Leguminosae show significant variation in plastome size and structure. Significant structural variations have been reported in three studied legume subfamilies: in subfamily Papilionoideae, IR loss, expansion and contraction, multiple large inversions, gene and intron loss occur (Bruneau et al., 1990; Wojciechowski et al., 2000; Martin et al., 2014; Schwarz et al., 2015); in subfamily Caesalpinioideae, significant IR expansion and contraction, small inversion and gene loss occur (Dugas et al., 2015; Wang et al., 2017); in subfamily Cercidoideae, IR expansion and contraction, and several large inversions occur (Dugas et al., 2015; Wang et al., 2018). However, plastomes in three other subfamilies (Deterioideae, Duparquetioideae, and Dialioideae) have been unaddressed.
The Dialioideae has been recently recognized as a separate subfamily of Leguminosae (LPWG, 2017). Containing 17 genera and ca. 85 species, this subfamily is distributed in the tropical to warm regions in Africa, Australia, America, and Southern China (LPWG, 2017). Morphologically, the Dialioideae is highly diverse with multiple symmetries and widely varied numbers of floral organs (Zimmerman et al., 2017). Some key morphological traits displayed in all or nearly all species of the Dialioideae distinguish this group from other subfamilies, including the presence of highly branched thyroid (less racemes with distichous anthotaxy in Labichea Gaudich. ex DC., Petalostylis R.Br.; Irwin and Barneby, 1981; Tucker, 1998; LPWG, 2017), leaves mostly imparipinnate with alternate leaflets (rarely paripinnate with opposite leaflets in Eligmocarpus Capuroni and Poeppigia C.Presl; LPWG, 2017), commonly indehiscent drupaceous or samaroid fruit (Irwin and Barneby, 1981), and usually 1–2 seeds lacking pleurograms (LPWG, 2017). Some species from Dialioideae are economically important. For example, Dialium cochinchinense Pierre is a common fruit in Southeast Asia, and Distemonanthus benthamianus Baill. produces wood with high quality. Phylogenetic analysis has shown that Dialioideae is well supported as the sister to the clade consisting of subfamilies Caesalpinioideae and Papilionoideae (LPWG, 2017); however, some intergeneric relationships within this subfamily are poorly resolved or not well supported (Zimmerman et al., 2017). To our knowledge, no studies have examined plastome structural variation in Dialioideae.
In this study, we investigated plastome variation in nine species representing nine genera of Dialioideae. The major purpose of this study is to reveal plastome variation in Dialioideae and reconstruct a highly resolved backbone of this subfamily.
2. Materials and methods 2.1. Plastome data set selectionWe used nine plastomes from nine species representing nine genera of Dialioideae from Zhang et al. (2020). The nine species are Baudouinia sp., Dialium schlechteri Harms, Dicorynia paraensis Benth., Distemonanthus benthamianus, Labichea lanceolata Benth., Petalostylis labicheoides R.Br., Poeppigia procera C.Presl, Storckiella pancheri Baill., and Zenia insignis Chun (NCBI accessions are available in Appendix: Supplementary material 1). We rechecked the annotations of all plastomes. In addition, protein coding genes were double-checked by finding open reading frames using the Find ORFs function in Geneious v.9.0.2 (Kearse et al., 2012). We used the online tRNAscan-SE service (Schattner et al., 2005) to check the identification of tRNA genes.
2.2. Analysis of plastome contentThe GC content (%), the plastome size (bp), the length of IR (bp), SSC (bp), and LSC (bp), the gene number, and gene distributions of all annotated plastomes were examined in Geneious. Genome maps of all annotated plastomes were generated by the online Organellar Genome DRAW (Lohse et al., 2013), and enclosed as Appendix: Supplementary material 2.
2.3. Analysis of plastome structureThe gene names and other information at the junction between the IR region and the single copy region were obtained using Geneious v.9.14. To detect the presence of gene inversions, we aligned the plastomes of nine Dialioideae species through the progressiveMauve algorithm of Mauve v.2.3.1 (Darling et al., 2010) implemented in Geneious. The sequence identity plot of all nine sampled Dialioideae plastomes was analyzed by mVISTA with shuffle-LAGAN model (Frazer et al., 2004). The plastome of Cercis glabra Pamp. was downloaded from NCBI (NCBI accession: KY806281) as a reference for structural analysis.
To identify sequence divergence in Dialioideae plastomes, 81 coding regions and 124 noncoding regions of 10 plastomes were extracted and aligned using MAFFT v.7.271 (Katoh and Standley, 2013). The variable sites and parsimony-informative sites in coding regions and noncoding regions were identified by PAUP v.4.0 (Swofford, 2002). Nucleotide diversity (π) of each region was calculated by a python script "nucleotide_freqs_by_site" (https://github.com/KatyBrown/).
2.4. Phylogenetic analysisTo reconstruct the phylogenetic relationships among Dialioideae, we complemented our data set of nine Dialioideae species with Duparquetia orchidacea Baill (NCBI accession: MN709829) of Duparquetioideae and Cercis glabra of Cercidoideae as outgroups. Four rRNA and 30 tRNA genes were not used in phylogenetic analysis due to limited parsimony-informative sites. Following the methods of Zhang et al. (2020), the 77 protein-coding genes (CDS) were extracted and aligned to generate alignment using MAFFT with LINSI algorithm.
The Maximum Likelihood (ML) tree was reconstructed by RAxML-HPC2 v.8.1 on XSEDE (Stamatakis, 2014) with the GTR+Γ substitution model and 1000 replicates of rapid bootstrap. The phylogenetic tree was visualized by FigTree v.1.4.3 (http://tree.bio.ed.ac.uk/).
3. Results 3.1. Plastome organizationResults of the sampling information and NCBI accessions of nine Dialioideae plastomes in this study are provided in Appendix: Supplementary material 1. The mean coverage of the nine Dialioideae plastomes ranged between 132.5 × (Dicorynia paraensis Benth.) and 2211.5 × (Labichea lanceolata) (Table 1). All plastomes show the typical quadripartite structure, a similar gene content and order. Among the nine plastomes, the smallest one in length is 154, 124 bp (Labichea lanceolata), and the largest in length is 165, 973 bp (Poeppigia procera). The lengths of the LSC, SSC, and IR range from 84, 755 bp (D. benthamianus) to 91, 963 bp (P. procera), 18, 048 bp (Labichea lanceolata) to 20, 188 bp (D. benthamianus), and 25, 640 bp (L. lanceolata) to 28, 326 bp (D. benthamianus), respectively (Table 1). The average GC content of the plastomes of nine species in Dialioideae is 36.19% (Supplementary material 2; Table 1). These nine plastomes have 129–132 genes, including 83–88 protein-coding genes, 36–38 tRNA genes, and 8 rRNA genes (genes within IR regions were counted twice). Of these genes, 12 genes have introns, and 17 genes are located in IR regions (Fig. 1). The plastomes of all Dialioideae species examined, except D. benthamianus, have one copy each of rpl14, rps3 and rpl16, two copies of trnNGUU, and an extra pseudogene copy of the ycf1. Distemonanthus benthamianus has two copies each of rpl14, rps3 and rpl16, one copy of trnNGUU, and lacks a pseudogene copy of ycf1. There is only one copy of the rps19 gene in Dicorynia paraensis, but two copies of the rps19 gene in D. benthamianus and Poeppigia procera, while one functional copy and one pseudogenized copy in other species. Poeppigia procera has two copies of trnHGUG gene, while others only have one copy (Table 2).
| Species | Dialium schlechteri | Petalostylis labicheoides | Distemonanthus benthamianus | Dicorynia paraensis | Poeppigia procera | Baudouiniasp. | Labichea lanceolata | Storckiella pancheri | Zenia insignis | Cercis glabra |
| Genome size (bp) | 159, 190 | 157, 565 | 161, 595 | 160, 661 | 165, 973 | 159, 072 | 154, 124 | 157, 343 | 159, 784 | 159, 181 |
| LSC size (bp) | 87, 530 | 86, 491 | 84, 755 | 89, 541 | 91, 963 | 87, 966 | 84, 796 | 86, 578 | 88, 915 | 88, 240 |
| Coverage (×) | 1806.4 | 906.6 | 402.6 | 132.5 | 2021.0 | 611.0 | 2211.5 | 1363.6 | 583.6 | – |
| IR (A/B) size (bp) | 26, 565 | 25, 900 | 28, 326 | 26, 070 | 27, 659 | 26, 235 | 25, 640 | 26, 058 | 26, 086 | 25, 625 |
| SSC size (bp) | 18, 530 | 19, 274 | 20, 188 | 18, 980 | 18, 692 | 18, 636 | 18, 048 | 18, 649 | 18, 697 | 19, 691 |
| GC content (%) | 36.2 | 36.6 | 35.9 | 36.0 | 35.2 | 36.2 | 36.8 | 36.5 | 36.3 | 36.2 |
| CDS | 83 | 83 | 88 | 83 | 84 | 83 | 83 | 83 | 83 | 83 |
| Total gene | 130 | 130 | 132 | 129 | 131 | 130 | 130 | 130 | 130 | 130 |
| rRNA gene | 8 | 8 | 8 | 8 | 8 | 8 | 8 | 8 | 8 | 8 |
| tRNA gene | 37 | 37 | 36 | 37 | 38 | 37 | 37 | 37 | 37 | 37 |
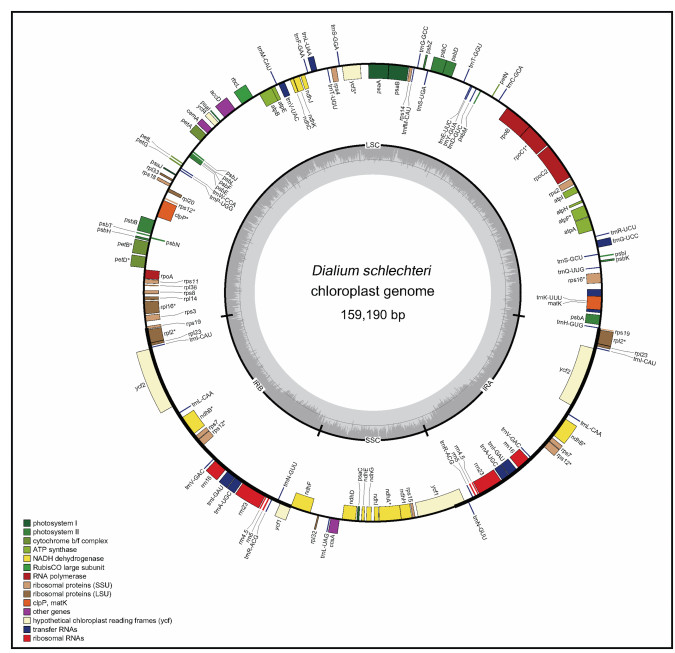
|
| Fig. 1 Gene map of the plastome of Dialium schlechteri. Boxes inside and outside the circle refer to the forward genes and reverse genes. Different types of genes are colored in different colors. Genes containing introns are marked by *. |
| Gene group | Gene name |
| Large subunit of ribosomal proteins | *rpl2, ^rpl14, ^rpl16, rpl20, *rpl23, rpl32, rpl33, rpl36 |
| Small subunit of ribosomal proteins | rps2, ^rps3, rps4, *rps7, rps8, rps11, *rps12, rps14, rps15, rps16, rps18, ^#Ψrps19 |
| Ribosomal RNA genes | *rrn4.5, *rrn5, *rrn16, *rrn23 |
| Transfer RNA genes | *trnA-UGC, trnC-GCA, trnD-GUC, trnE-UUC, trnF-GAA, trnfM-CAU, trnG-GCC, trnG-UCC, #trnH-GUG, *trnI-CAU, *trnI-GAU, trnK-UUU, *trnL-CAA, trnL-UAA, trnL-UAG, trnM-CAU, *trnN-GUU, trnP-UGG, trnQ-UUG, *trnR-ACG, trnR-UCU, trnS-GCU, trnS-GGA, trnS-UGA, trnT-GGU, trnT-UGU, *trnV-GAC, trnV-UAC, trnW-CCA, trnY-GUA |
| DNA-dependent RNA polymerase | rpoA, rpoB, rpoC1, rpoC2 |
| Photosystem Ⅰ | psaA, psaB, psaC, psaI, psaJ |
| Photosystem Ⅱ | petA, petB, petD, petG, petL, petN |
| Cytochrome b/f complex | psbA, psbB, psbC, psbD, psbE, psbF, psbH, psbI, psbJ, psbK, psbL, psbM, psbN, psbT, psbZ |
| ATP synthase | atpA, atpB, atpE, atpF, atpH, atpI |
| Maturase K | matK |
| Envelope membrane proteins | cemA |
| subunit of acetyl-CoA | accD |
| RubisCo large subunit | Rbcl |
| Hypothetical reading frames | Ψycf1, *ycf2, ycf3, ycf4 |
| Protease | clpP |
| NADH dehydrogenase | ndhA, *ndhB, ndhC, ndhD, ndhE, ndhF, ndhG, ndhH, ndhI, ndhJ, ndhK |
| c-type cytochrome | ccsA |
Six of nine Dialioideae plastomes (D. schlechteri, P. labicheoides, Baudouinia sp., Labichea lanceolata, S. pancheri and Z. insignis) have canonical IR regions (ranging from 25, 640 bp in Labichea lanceolata to 26, 565 bp in D. schlechteri; Table 1). Within these six plastomes, the LSC/IRB junction (JLB) is located in rps19, resulting in the duplication of the 3′ end of this gene (85 bp in Z. insignis, and 74 bp in the other four species); the SSC/IRA junction (JSA) is located in ycf1, moving 520–1221 bp of the 3′ end of this gene into the IR; the SSC/IRB junction (JSB) is located between the duplicated 3′ end of ycf1 and ndhF; and the LSC/IRA junction (JLA) is located between the duplicated 3′ end of rps19 and trnHGUG. However, the IR size and boundaries show significant variation in D. benthamianus, P. procera and D. paraensis (Fig. 2). The IR of D. benthamianus has expanded its JLB into LSC to include four more complete genes (rps19, rps3, rpl16, rpl14), its JSB into SSC to include 19 bp of the 5′ end of ndhF, and has contracted its JSA to release the complete ycf1 and trnNGUU into the SSC (Fig. 2). The IR of P. procera has expanded its JLB into the LSC to include rps19, and expanded its JLA into the LSC to include trnHGUG. The IR of D. paraensis has experienced contraction out of the LSC, moving the whole sequence of rps19 into the LSC.
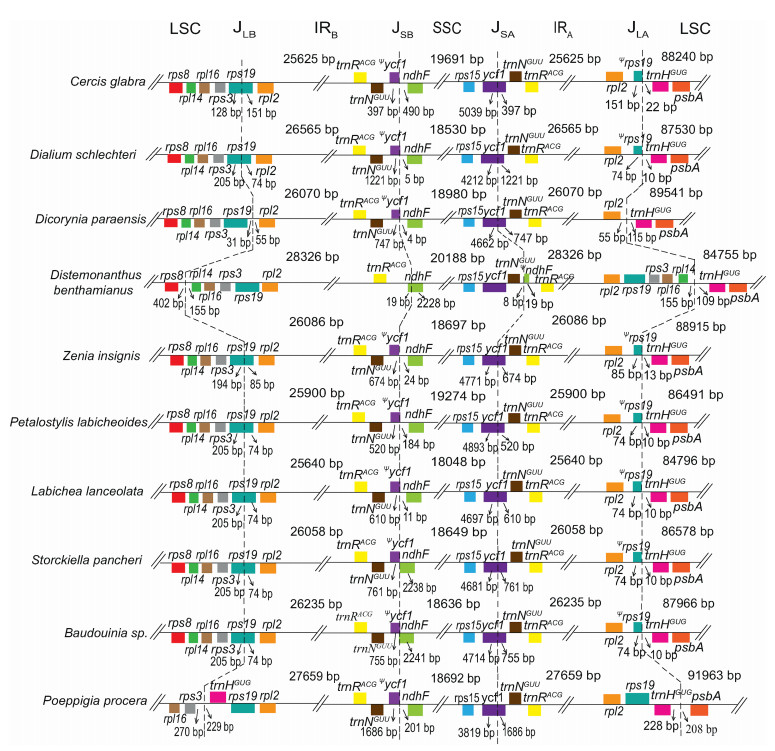
|
| Fig. 2 IR/SC junctions in Dialioideae. JLB, JSB, JSA, and JLA refer to the junctions of LSC/IRB, SSC/IRB, SSC/IRA, and LSC/IRA. The boxes above and below the line refer to the forward and reverse genes. The tree on the left is built by RAxML-HPC2 on XSEDE. Pseudogenes are marked by Ψ. |
The number of variable sites and parsimony-informative sites from 81 coding regions and 124 noncoding regions were collected. Among coding regions, the percentage of variable sites ranges from 0 (rrn5 gene) to 24.82% (ycf1 gene), and the percentage of parsimony-informative sites ranges from 0 (petN, psbL, petL, rrn4.5 and rrn5 genes) to 6.83% (ycf1 gene) (Fig. 3a and Appendix: Supplementary material 3). Among noncoding regions, the percentage of variable sites ranges from 0.05 (trnNGUU-ycf1) to 48.24% (psbK-trnQUUG), and the percentage of parsimony-informative sites ranges from 0 (five intergenic regions including ndhK-ndhC, rpl23-trnICAU, trnICAU-ycf2, rrn16-trnVGAC and rrn23-rrn4.5) to 22.14% (accD-rbcL) (Fig. 3b and Appendix: Supplementary material 3). Among the whole plastome, coding regions are more conserved than noncoding regions, and IR regions are more conserved than LSC and SSC regions (Figs. 3c and 4). The mean value of the percentage of variable sites from coding regions is 7.81%, which is around 40% that from noncoding regions (19.60%). Similarly, the mean value of the percentage of parsimony-informative sites from noncoding regions is 5.92%, which is around 2.65 times that from coding regions (2.23%). The mean percentage of variable sites and parsimony-informative sites in the IR region are 3.66% and 0.95%, which are lower than that in the LSC region (16.90% for variable sites and 5.13% for parsimony-informative sites) and the SSC region (18.41% for variable sites and 5.14% for parsimony-informative sites) (Appendix: Supplementary material 4).
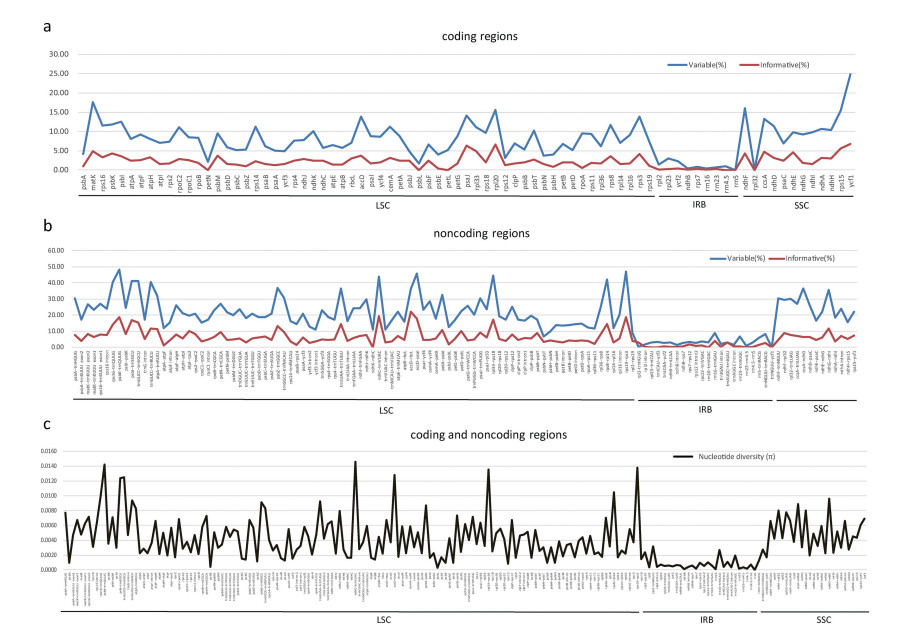
|
| Fig. 3 The percentages of variable sites and parsimony informative sites, and nucleotide diversity (π) across coding regions and noncoding regions (a The percentages of variable sites and parsimony informative sites in coding regions, b The percentages of variable sites and parsimony informative sites in noncoding regions, c nucleotide diversity in coding and noncoding regions). |
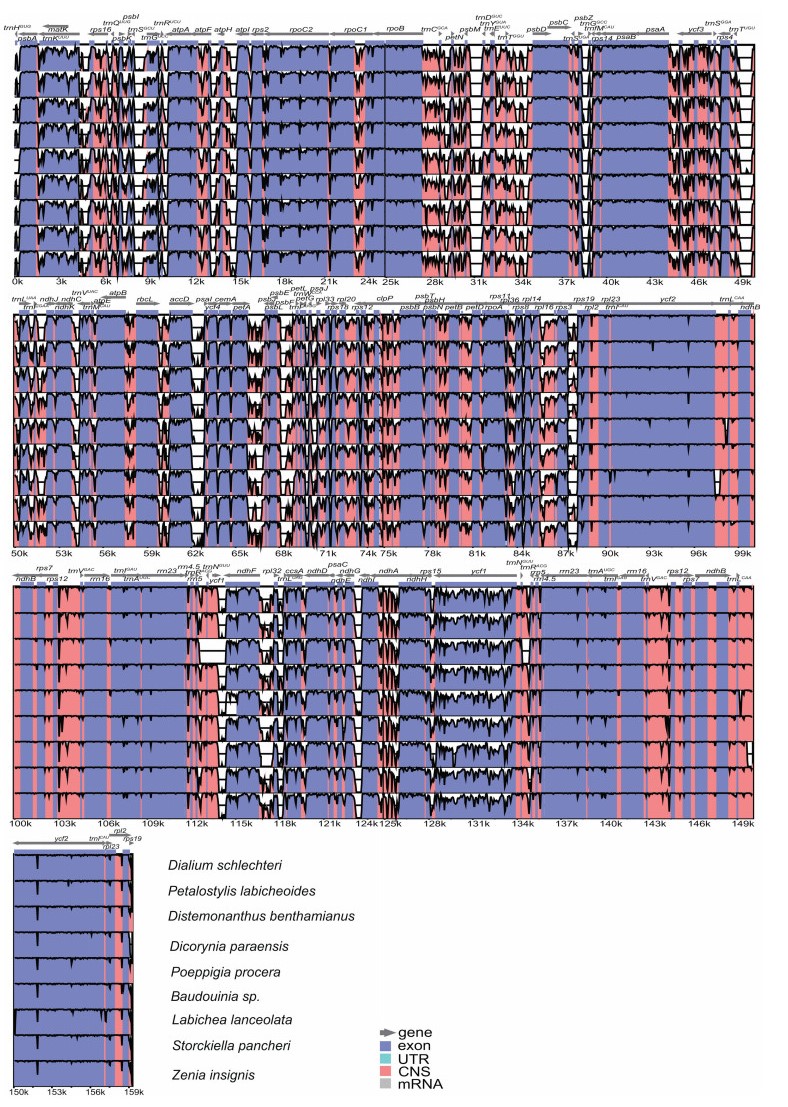
|
| Fig. 4 Sequence alignment of complete plastome sequences of Dialioideae and outgroup samples compared in this study using the mVISTA program. The vertical scale represents the percent of identity ranging from 50% to 100%. Cercis glabra is used as a reference. |
Phylogenetic analysis based on 77 protein-coding gene sequences fully resolves phylogenetic relationships among sampled species with strong bootstrap support (BS = 100%) for all nodes except a moderately supported (BS = 63%) sister relationship between Petalostylis labicheoides and Labichea lanceolata (Fig. 5). The monophyly of Dialioideae is strongly supported (BS = 100%). Poeppigia procera and Baudouinia sp. are supported as the successive sisters to other Dialioideae, which is further divided into two clades (clade A and clade B).
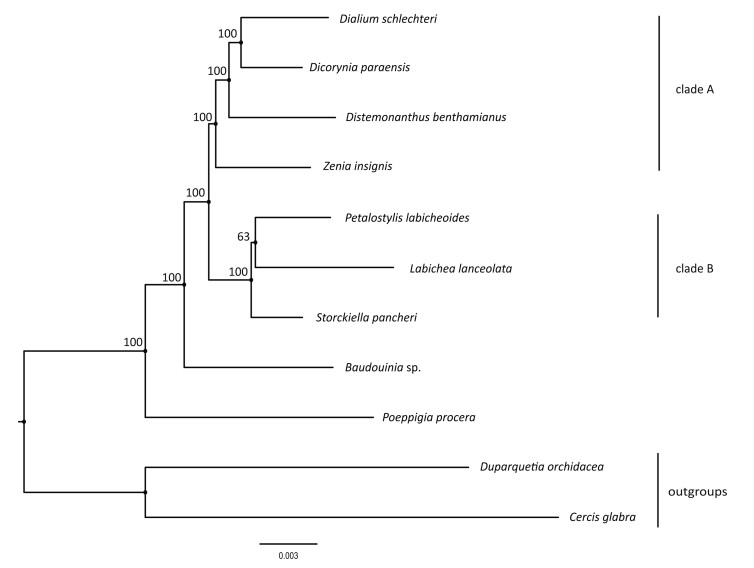
|
| Fig. 5 Phylogeny of Dialioideae species inferred from plastome protein coding genes using the maximum likelihood (ML) method. The ML tree shows bootstrap values on each node. |
Plastome of three legume subfamilies (Papilionoideae, Caesalpinioideae and Cercidoideae) have experienced significant structural variations. In Papilionoideae, plastome structural variation includes an IR loss in one large clade (Wojciechowski et al., 2000), multiple large inversions (e.g., a 78-kb inversion in tribe Phaseoleae, a 36-kb inversion in Lupinus L., a 39-kb inversion in Robinia L.) (Bruneau et al., 1990; Martin et al., 2014; Schwarz et al., 2015), multiple losses of introns and genes (Downie et al., 1991; Jansen et al., 2008; Martin et al., 2014), and multiple losses of intergenic spacer sequences (Schwarz et al., 2015). In Cercidoideae, plastome variations include large inversions (38-kb, 24-kb and 7.5-kb), large IR expansions and contractions (Kim and Cullis, 2017; Wang et al., 2018). In Caesalpinioideae, multiple IR expansions and contractions, and the loss of rpl22 have been observed in some species (Dugas et al., 2015; Wang et al., 2017). However, aside for some size variation, plastomes of Dialioideae have a relatively conserved structure. This study shows that not all legume lineages have experienced significant plastome structural variation.
The reduction of plastome size is mainly caused by IR contraction or loss, gene loss, intron loss, or intergenic spacer loss (Jansen et al., 2008; Dugas et al., 2015; Schwarz et al., 2015). In contrast, the increase of plastome size is usually caused by IR expansion and gene duplication (Dugas et al., 2015; Wang et al., 2018). The plastome size of nine Dialioideae species varies from 154, 123 to 165, 973 bp, and gene number varies from 129 to 132. These differences are mainly caused by IR expansion and contraction in three species. The IR of Distemonanthus benthamianus has experienced two expansions to include four genes and part of ndhF, and one contraction to move the whole ycf1 and trnNGUU sequences into the SSC. The IR of Poeppigia procera has experienced two expansions to include two genes, and the IR of Dicorynia paraensis has experienced one contraction that caused the partial loss of rps19.
Plastome sequences have been widely used to infer relationships at all taxonomic levels from deep relationships of land plants, through relationships among orders, families or genera, to relationships among species or populations (see Shaw et al., 2014). RbcL and matK have been proposed as core barcodes for land plants (CBOL Plant Working Group, 2009), and psbA–trnH as a complementary barcode (China Plant BOL Group, 2011). Ten plastid noncoding loci (5′rps16-trnQUUG, ndhC-trnVUAC, ndhF-rpl32, trnT GGU-psbD, psbE-petL, petA-psbJ, rpl32-trnLUAG, rpl16 intron, ndhA intron and rpoB-trnCGCA) have been consistently considered the most variable regions across angiosperm lineages (Shaw et al., 2014). Similarly, ndhC-trnVUAC (0.0146) and 5′rps16-trnQUUG (0.01) are two highly variable regions with high nucleotide diversity. Aside from these sequences, we found that the seven most informative regions for Dialioideae are psbK-trnQUUG (0.0142), rps19-rps3 (0.0138), rpl33-rps18 (0.0135), accD-psaI (0.0128), trnGUCC-trnSGCU (0.0125), psbI-trnSGCU (0.0124) (Fig. 3c and Appendix: Supplementary material 3). In addition, the ycf1 gene had the highest nucleotide diversity among all protein coding genes. It is worth determining whether these sites can be used as potential molecular markers to clarify close relationships between species and populations in Dialioideae.
Previous work suggested that the subfamily Dialioideae was strongly supported (LPWG, 2017; Zhang et al., 2020). Our study confirms strong support for this subfamily. However, intergeneric relationships within Dialioideae have been poorly resolved or not well supported in previous studies that relied on a few plastid loci and/or morphological characters (Bruneau et al., 2001; Herendeen et al., 2003; Bruneau et al., 2008; LPWG, 2017; Zimmerman et al., 2017). Our study fully resolves most intergeneric relationships with strong support (BS = 100%) except those among Labichea, Petalostylis and Storchiella. Previous studies have also failed to resolve the relationships among Labichea, Petalostylis and Storchiella. Bruneau et al. (2001) reconstructed a phylogeny with a weakly supported relationship of (Storchiella, Petalostylis); however, Zimmerman et al. (2017) supported a relationship of (Storchiella, (Labichea, Petalostylis)), among them, the bootstrap support for a relationship (Labichea, Petalostylis) was less than 85%. Our analysis, which supports the same topology as Zimmerman et al. (2017), indicates that the relationship between Labichea and Petalostylis is weakly supported. Although further studies are needed to clarify the relationships among Labichea, Storchiella and Petalostylis, this study shows that chloroplast phylogenomics offers an efficient approach to build a robust tree of Dialioideae.
Author contributionsT-SY, RZ and H-RB designed this research, RZ checked plastome annotation, reconstructed the phylogenetic tree; H-RB and OO conducted other analyses, H-RB and T-SY wrote the manuscript, H-RB, RZ and T-SY revised the manuscript.
Declaration of Competing InterestThe author declares no conflict of interest.
AcknowledgmentsThis study was supported by grants from the Large-scale Scientific Facilities of Chinese Academy of Sciences (No. 2017-LSF-GBOWS-02), the Strategic Priority Research Program of Chinese Academy of Sciences (XDB31010000), the National Natural Science Foundation of China [key international (regional) cooperative research project No. 31720103903]. We are grateful to Jian-Jun Jin, Shu-Dong Zhang and Xiao-Jian Qu for helpful discussions during the data analysis.
Appendix A. Supplementary dataSupplementary data to this article can be found online at https://doi.org/10.1016/j.pld.2020.06.008.
Bruneau A., Doyle J.J., Palmer J.D., 1990. A chloroplast DNA inversion as a subtribal character in the Phaseoleae (Leguminosae). Syst. Bot, 15: 378-386. DOI:10.1600/036364409787602221 |
Bruneau A., Forest F., Herendeen P.S., et al, 2001. Phylogenetic relationships in the Caesalpinioideae (Leguminosae) as inferred from chloroplast trnL intron sequences. Syst. Bot, 26: 487-514. DOI:10.1043/0363-6445-26.3.487 |
Bruneau A., Mercure M., Lewis G.P., et al, 2008. Phylogenetic patterns and diversification in the caesalpinioid legumes. Botany, 86: 697-718. DOI:10.1139/B08-058 |
CBOL Plant Working Group, 2009. A DNA barcode for land plants. Proc. Natl. Acad.Sci. U. S. A, 106: 12794-12797. DOI:10.1073/pnas.0905845106 |
China Plant BOL Group, 2011. Comparative analysis of a large dataset indicates that internal transcribed spacer (ITS) should be incorporated into the core barcode for seed plants. Proc. Natl. Acad. Sci. U. S. A, 108: 19641-19646. DOI:10.1073/pnas.1104551108 |
Darling A.E., Mau B., Perna N.T., 2010. Progressivemauve: multiple genome alignment with gene gain, loss and rearrangement. PloS One, 5: e1147. DOI:10.1371/journal.pone.0011147 |
Downie S.R., Olmstead R.G., Zurawski G., et al, 1991. Six independent losses of the chloroplast DNA rpl2 intron in dicotyledons: molecular and phylogenetic implications. Evolution, 45: 1245-1259. DOI:10.1111/j.1558-5646.1991.tb04390.x |
Dugas D.V., Hernandez D., Koenen E.J.M., et al, 2015. Mimosoid legume plastome evolution: IR expansion, tandem repeat expansions, and accelerated rate of evolution in clpP. Sci. Rep, 5: 16958. DOI:10.1038/srep16958 |
Frazer K.A., Pachter L., Poliakov A., et al, 2004. Vista: computational tools for comparative genomics. Nucleic Acids Res, 32: W273-W279. DOI:10.1093/nar/gkh458 |
Herendeen, P.S., Bruneau, A., Lewis, G.P., 2003. Phylogenetic relationships in Caesalpinioid legumes: a preliminary analysis based on morphological and molecular data. In: Klitgaard, B.B. (Ed. ), Advances in Legume Systematics 10. Royal Botanic Gardens, Kew, pp. 37-62.
|
Irwin, H.S., Barneby, R.C., 1981. Cassieae. In: Polhill, R.M., Raven, P.H. (Eds. ), Advances in Legume Systematics 1. Royal Botanic Gardens, Kew, pp. 97-106.
|
Jansen R.K., Wojciechowski M.F., Sanniyasi E., et al, 2008. Complete plastome sequence of the chickpea (Cicer arietinum) and the phylogenetic distribution of rps 12 and clpP intron losses among legumes (Leguminosae). Mol. Phylogenet.Evol, 48: 1204-1217. DOI:10.1016/j.ympev.2008.06.013 |
Jansen, R.K., 2012. Plastomes of seed plants. In: Bock, R., Knoop, V. (Eds. ), Advances in Photosynthesis and Respiration. Springer, Dordrecht Advances, pp. 103-126.
|
Katoh K., Standley D.M., 2013. MAFFT multiple sequence alignment software version 7:improvements in performance and usability. Mol. Biol. Evol, 30: 772-780. DOI:10.1093/molbev/mst010 |
Kearse M., Moir R., Wilson A., et al, 2012. Geneious basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics, 28: 1647-1649. DOI:10.1093/bioinformatics/bts199 |
Kim Y., Cullis C., 2017. A novel inversion in the chloroplast genome of marama (Tylosema esculentum). J. Exp. Bot, 68: 2065-2072. DOI:10.1093/jxb/erw500 |
Lohse M., Drechsel O., Kahlau S., et al, 2013. OrganellarGenomeDRAW-a suite of tools for generating physical maps of plastid and mitochondrial genomes and visualizing expression data sets. Nucleic Acids Res, 41: W575-W581. DOI:10.1093/nar/gkt289 |
LPWG[LegumePhylogeny Working Group], 2017. A new subfamily classification of the Leguminosae based on a taxonomically comprehensive phylogeny. Taxon, 66: 44-77. DOI:10.12705/661.3 |
Martin G.E., Rousseau-Gueutin M., Cordonnier S., et al, 2014. The first complete plastome of the Genistoid legume Lupinus luteus: evidence for a novel major lineage-specific rearrangement and new insights regarding plastome evolution in the legume family. Ann. Bot, 113: 1197-1210. DOI:10.1093/aob/mcu050 |
Schattner P., Brooks A.N., Lowe T.M., 2005. The tRNAscan-SE, snoscan and snoGPS web servers for the detection of tRNAs and snoRNAs. Nucleic Acids Res, 33: W686-W689. DOI:10.1093/nar/gki366 |
Shaw J., Shafer H.L., Leonard O.R., et al, 2014. Chloroplast DNA sequence utility for the lowest phylogenetic and phylogeographic inferences in angiosperms: the tortoise and the hare Ⅳ. Am. J. Bot, 101: 1987-2004. DOI:10.3732/ajb.1400398 |
Schwarz E.N., Ruhlman T.A., Sabir J.S.M., et al, 2015. Plastome sequences of legumes reveal parallel inversions and multiple losses of rps 16 in papilionoids. J. Syst. Evol, 53: 458-468. DOI:10.1111/jse.12179 |
Stamatakis A., 2014. RAxML version 8:a tool for phylogenetic analysis and postanalysis of large phylogenies. Bioinformatics, 30: 1312-1313. DOI:10.1093/bioinformatics/btu033 |
Swofford, D., 2002. PAUP*: Phylogenetic Analysis Using Parsimony (*and Other Methods), Version 4.0b10. Sinauer Associates.
|
Tucker S.C., 1998. Floral ontogeny in legume genera Petalostylis, Labichea, and Dialium (Caesalpinioideae: cassieae), a series in floral reduction. Am. J. Bot, 85: 184-208. DOI:10.2307/2446307 |
Wang Y.-H., Qu X.-J., Chen S.-Y., et al, 2017. Plastomes of Mimosoideae: structural and size variation, sequence divergence, and phylogenetic implication. Tree Genet. Genomes, 13: 41. DOI:10.1007/s11295-017-1124-1 |
Wang Y.-H., Wicke S., Wang H., et al, 2018. Plastome evolution in the earlydiverging legume subfamily Cercidoideae (Fabaceae). Front. Plant Sci, 9: 138. DOI:10.3389/fpls.2018.00138 |
Wicke S., Schneeweiss G.M., dePamphilis C.W., et al, 2011. The evolution of the plastid chromosome in land plants: gene content, gene order, gene function. Plant Mol. Biol, 76: 273-297. DOI:10.1007/s11103-011-9762-4 |
Wojciechowski, M.F., Sanderson, M.J., Steele, K.P., et al., 2000. Molecular phylogeny of the "temperate herbaceous tribes" of papilionoid legumes: a supertree approach. In: Herendeen, P.S., Bruneau, A. (Eds. ), Advances in Legume Systematics 9. Royal Botanic Gardens, Kew, pp. 277-298.
|
Zhang R., Wang Y.-H., Jin J.-J., et al, 2020. Exploration of plastid phylogenomic conflict yields new insights into the deep relationships of Leguminosae. Syst.Biol, 69: 613-622. DOI:10.1093/sysbio/syaa013 |
Zimmerman E., Herendeen P.S., Lewis G.P., et al, 2017. Floral evolution and phylogeny of the Dialioideae, a diverse subfamily of tropical legumes. Am. J. Bot, 104: 1019-1041. DOI:10.3732/ajb.1600436 |



