b. University of Chinese Academy of Sciences, Beijing, 100049, China;
c. College of Life Sciences, Guizhou University, Guiyang, Guizhou, 550025, China
Salvia L., which includes about 1000 species, is the largest genus of Lamiaceae (Hu et al., 2018). Salvia is widely distributed in the world, mainly in Mesoamerica, South America, East Asia, the Mediterranean region, and south-western Asia (Walker and Sytsma, 2006; Wei et al., 2015). Most species of Salvia have important medicinal, ornamental, and economic values (e.g., Salvia miltiorrhiza Bung., Salvia splendens Ker Gawl., and Salvia hispanica L.) (Ge et al., 2014; Ali et al., 2012; Wang, 2010). Salvia is well known for its highly specialized flower structure and highly diverse staminal morphology which forms the staminal lever mechanism (Claßen-Bockhoff et al., 2008).
Recent molecular phylogenetic studies have suggested that traditionally defined Salvia is not monophyletic (Drew et al., 2017; Fragoso-Martínez et al., 2018; Hu et al., 2018; Jenks et al., 2012; Kriebel et al., 2019; Li et al., 2013; Walker et al., 2004; Will and Claßen-Bockhoff 2014; Will and Classen-Bockhoff, 2017). Some researchers (Drew et al., 2017; Hu et al., 2018) have proposed that Salvia should be expanded to include five additional genera (Dorystaechas Boiss. & Heldr. ex Benth., Meriandra Benth., Perovskia Karel., Rosmarinus L. and Zhumeria Rech. & Wendelbo). Will and Classen-Bockhoff (2017) have recommended dividing the genus of Salvia into six genera. Here, we adopt a broad definition of Salvia following the justification of Drew et al. (2017).
Recently, sequencing technology and analytical methods have developed rapidly, and some plastid DNA fragments, such as psbA–trnH, matK, rbcL, trnL–trnF, ycf1, and nuclear ribosomal Internal Transcribed Spacers (nrITS), have been utilized to study the phylogeny of Salvia (Chen et al., 2009). Drew et al. (2017) utilized two low-copy nuclear gene regions (PPR–AT3G09060 and GBSSI) and four plastid markers (psbA–trnH, trnL–trnF, ycf1, and ycf1–rps15) to infer the phylogenetic relationships of Salvia. Their results primarily confirmed that Salvia in the traditional sense was non-monophyletic, embedding five additional genera (Dorystaechas, Meriandra, Perovskia, Rosmarinus and Zhumeria) based on plastid and nuclear DNA data. Using nrITS and plastid marker rpl32–trnL, Will and Classen-Bockhoff (2017) found that Salvia species can be divided into four lineages, with the above-mentioned five genera clustered within them. Meanwhile, Fragoso-Martínez et al. (2017) used an anchored hybrid enrichment method to resolve relationships among subgenus Calosphace. In addition, Hu et al. (2018) used two nuclear markers (nrITS and ETS) and four plastid DNA markers (psbA–trnH, ycf1–rps15, trnL–trnF and rbcL) to reconstruct the phylogenetic analysis of East Asian Salvia. Using the same approach, Kriebel et al. (2019) provided a well-resolved backbone phylogeny of Salvia. However, previous analyses have all been based on DNA markers, whereas few genomic data sets have been used to construct a phylogeny of Salvia.
Altogether, Salvia is divided into 11 subgenera based on molecular and morphological data (Drew et al., 2017; Hu et al., 2018; Kriebel et al., 2019; Will and Classen-Bockhoff, 2017). East Asia is one of the distribution centers of Salvia, with approx. 100 species in total, more than 80% of which are in China (Hu et al., 2018). Based on phylogenetic analysis focused on Salvia in East Asia, Hu et al. (2018) established a subgenus, subg. Glutinaria, to accommodate all species of Salvia native to East Asia. There are also many important medical plants in this subgenus, such as S. miltiorrhiza, S. cavaleriei Levl and S. yunnanensis C. H. Wright (Qian et al., 2002; Wang, 2010; Zhang and Li, 1994). Few early molecular phylogenetic studies have focused on species currently placed in Salvia subg. Glutinaria. Takano and Okada (2011) used three DNA markers (rbcL, trnL–trnF and nrITS) to report that 11 Japanese Salvia species were monophyletic. Using four DNA markers (psbA–trnH, rbcL, matK and nrITS), Li et al. (2013) demonstrated that 41 species of Salvia from China and Japan formed a clade. Hu et al. (2018) used 93 taxa to reconstruct the phylogenetic relationships of Salvia subg. Glutinaria and recognized eight sections within this subgenus. However, interspecific relationships of Salvia subg. Glutinaria remain unresolved.
The chloroplast genome, or plastome, has long been used in plant phylogenetics. Several recent studies have found that using plastomes can powerfully resolve the phylogenetic relationship of plants at different taxonomic levels (Lin et al., 2012; Wang et al., 2016; Yang et al., 2013a, 2013b). Plastomes are relatively stable, easy to obtain, and contain a large amount of information with a suitable mutation rate (Tian and Li, 2002). The plastome contains four extremely evolutionarily conserved parts: a pair of inverted repeat regions (IRa and IRb), a large single-copy region (LSC), and a small single-copy region (SSC) in most land plants. Structural variation and the presence/absence of genes in these genomes can also be used for phylogenetic inference.
In total, 53 Lamiaceae plastomes have been sequenced and are available for public use (NCBI GenBank database, accessed on 31 Jan 2020). The structure and organization of Lamiaceae plastomes is conserved and stable, and consists of the quadripartite plastome structure of most angiosperms. Lamiaceae plastomes possess between 80 and 81 unique protein-coding genes, range in length from 149, 736 to 155, 293 bp, and have a GC content between 37.8 and 38.7% (Lindqvist and Albert, 2002). However, there are only seven plastomes from Salvia publicly available. Hence, the plastome characterization of Salvia is still far from adequate and few phylogenetic trees of Salvia have been reconstructed using plastomes in previous phylogenetic relationship of Salvia, particularly in the East Asian subg. Glutinaria.
Here, we analyzed Salvia phylogeny using plastome sequence data. The main objectives of this research are to (1) investigate structural and compositional variation of plastomes in East Asian Salvia species; (2) estimate the feasibility of using plastome data for phylogenetic reconstruction of the Salvia subg. Glutinaria; and (3) find the optimal DNA regions to better construct the phylogenetic tree of Salvia at large.
2. Materials and methods 2.1. Plant materialsWe sampled a total of twenty-one Salvia accessions from nineteen different species. Fresh green leaves of fourteen Salvia species and two Melissa L. were acquired from the field and sequenced. Two species of Melissa were selected as outgroup based on previous phylogenetic studies (Hu et al., 2018; Kriebel et al., 2019). Voucher specimens were deposited at the herbarium of the Kunming Institute of the Botany, Chinese Academy of Sciences (KUN) (for details see Table 1). Five Salvia plastomes (S. miltiorrhiza, S. rosmarinus Spenn., S. officinalis L., S. chanryoenica Nakai and S. bulleyana Diels) were downloaded from GenBank for analyses. Previously reported data for two Salvia species were not used because these species may have been misidentified (Salvia japonica Thunb (He et al., 2017).) or were re-sequenced in this study (Salvia przewalskii Maxim.). This data set represents all four subgenera of Salvia and each of the four sections of subg. Glutinaria.
| Taxon | Voucher | Accession numbers |
| Ingroup | ||
| Salvia brachyloma Stib. | 10CS2189 | MT634136 |
| S. bulleyana Diels | – | NC_041092 |
| S. castanea Diels f. castanea | 11CS3534 | MT634150 |
| S. castanea Diels f. tomentosa | LiuJQ–08XZ–091 | MT634141 |
| S. cavaleriei Levl. | 10CS1700 | MT634139 |
| S. chanryoenica Nakai | – | NC_040121 |
| S. cyclostegia Stib. | HP8813 | MT634144 |
| S. flava Forrest ex Diels | 11CS3465 | MT634140 |
| S. mairei Levl. | HP8366 | MT634143 |
| S. mekongensis Stib. | HGW–00244 | MT634146 |
| S. miltiorrhiza Bunge | – | NC_020431 |
| S. officinalis L. | – | NC_038165 |
| S. plectranthoides Griff. | 10CS2117 | MT634138 |
| S. przewalskii Maxim. | ZhouZK–07ZX–0342 | MT634135 |
| S. rosmarinus Spenn. 1 | 13CS6707 | MT634145 |
| S. rosmarinus Spenn. 2 | – | NC_027259 |
| S. sp. | LJBG0615 | MT634149 |
| S. subpalmatinervis Stib. | YangQE1866 | MT634137 |
| S. tiliifolia Vahl | YNS0517 | MT634134 |
| S. umbratica Hance | 10CS2479 | MT634142 |
| S. yunnanensis C. H. Wright | 10CS1936 | MT634133 |
| Outgroup | ||
| Melissa axillaris (Benth.) Bakh. f. | GLGE12054 | MT634147 |
| M. yunnanensis C. Y. Wu et Y. C. Huang | 11CS3711 | MT634148 |
Chloroplast DNA was extracted following the CTAB method (Doyle, 1987). After purification, we fragmented 5 mg DNA to build short-insert libraries, following the manufacturer's instructions (Illumina). We used tags to index DNA from different individuals and pooled samples before sequencing in Illumina's Genomic Analyzer. Low-quality reads were discarded using the NGS QC Toolkit (Patel and Jain, 2012). Sequence reads were filtered from the raw sequence reads through comparison to published sequences of S. miltiorrhiza (NC_020431) (Qian et al., 2013). The remaining sequences were assembled into complete plastomes de novo using GetOrganelle (Jin et al., 2018). Complete plastomes were visualized in Bandage v.8.1. (Wick et al., 2015). Finally, the order of the complete plastomes was determined by comparison to the reference genome S. miltiorrhiza.
2.3. Chloroplast genome annotationPlastomes were annotated using Plastid Genome Annotator (PGA) genomes and manually revised (Qu et al., 2019). The tRNA boundaries were then identified with tRNAscan-SE (v.1.3.1) (Lowe and Chan, 2016). Salvia plastome gene maps were drawn with the OrganellarGenomeDRAW tool (OGDRAW) (Wyman et al., 2004).
2.4. Comparative chloroplast genomic analysesTo investigate interspecies variation, whole plastome alignment of the 21 Salvia species and two species of Melissa was performed using the mVISTA program in Shuffle-LAGAN mode (A Frazer et al., 2004), with S. miltiorrhiza as the reference. IR expansion/contraction of these plastomes was also checked using online IRscope (Amiryousefi et al., 2018). The nucleotide diversity (Pi) of plastomes was estimated by DnaSP v.5.10 (Librado and Rozas, 2009) with step size set at 200 bp and window length set at 600 bp.
2.5. Plastid phylogenomic analysesIn this study, 23 plastome sequences, including those of two outgroups from subtribe Melissinae, were aligned for phylogenomic analysis. To examine the performance of different plastome regions in phylogenetic resolution, 10 matrices were created for phylogenetic analyses (Table 2). The first five matrices of plastid sequence data were as follows: (1) complete plastid DNA sequences containing only one IR (Plastomes); (2) protein-coding exons (CR); (3) introns and spacers (IS); (4) the large single-copy region and the small single-copy region (SCR); and (5) the inverted repeat region (IR). We created five additional matrices by eliminating rapidly evolving sites from each of the preceding data sets using Gblocks (Castresana, 2000) with default parameters. Plastome sequences were aligned using the Multiple Sequence Alignment Program (MAFFT v.5) and manually adjusted when necessary (Katoh et al., 2005). Ambiguously aligned regions were excluded.
| Data matrix | BI analysis model | Aligned (bp) | Constant sites (bp) | Variable sites (bp) | Parsimony-informative (bp) | |
| CR | GTR + I + G + F | 77, 918 | 66, 638 (85.52%) | 3037 | 1555 (2.00%) | |
| CR (Gblocks) | GTR + I + G + F | 69, 644 | 66, 618 (95.66%) | 3026 | 1552 (2.23%) | |
| IR | GTR + I + F | 28, 041 | 24, 031 (85.70%) | 555 | 119 (0.42%) | |
| IR (Gblocks) | GTR + I + F | 24, 540 | 23, 993 (97.77%) | 547 | 119 (0.48%) | |
| IS | GTR + I + G + F | 64, 040 | 40, 939 (63.93%) | 3826 | 1947 (3.04%) | |
| IS (Gblocks) | GTR + I + G + F | 43, 822 | 40, 155 (91.63%) | 3667 | 1849 (4.22%) | |
| Plastomes | GTR + I + G + F | 137, 226 | 113, 108 (82.42%) | 6973 | 3644 (2.66%) | |
| Plastomes (Gblocks) | GTR + I + G + F | 119, 172 | 112, 362 (94.29%) | 6810 | 3552 (2.89%) | |
| SCR | GTR + I + G + F | 108, 083 | 88, 326 (81.72%) | 6373 | 3505 (3.24%) | |
| SCR (Gblocks) | GTR + I + G + F | 93, 890 | 87, 659 (93.63%) | 6231 | 3425 (3.65%) | |
| CR: protein-coding exons; IR: inverted repeat region; IS: introns and spacers; Plastomes: the complete plastid DNA sequences only containing one IR; SCR: the large single-copy region and the small single-copy region; (Gblocks): indicates that rapidly evolving sites were eliminated by Gblocks. The percentages in 'Constant sites (bp)' and 'Parsimony-informative (bp)' represent 'Constant sites (bp)/Aligned (bp)' and 'Parsimony-informative (bp)/Aligned (bp)', respectively. | ||||||
Maximum likelihood (ML) analyses were conducted using RAxML v.8.2.9 (Stamatakis et al., 2005), under the general time-reversible model of nucleotide substitution with a gamma distribution of rate variation among sites (GTRGAMMA) with 1000 replicates and other default parameters.
Maximum parsimony (MP) analysis were performed with PAUP∗4.0b10 (Swofford, 2002). A 1000–replicate heuristic search was applied with random addition, and tree bisection-reconnection (TBR) branch swapping with the MUL-trees was used to search for MP trees. Parameter settings remained unchanged.
Bayesian inference (BI) analysis was carried out in MrBayes v.3.2.6 (Ronquist et al., 2012) in PhyloSuite (Zhang et al., 2020), using the ModelFinder (Kalyaanamoorthy et al., 2017) to select the best model based on Akaike information criterion (AIC). GTR + I + F was found to be the best-fit model for IR Regions and IR Regions (using Gblocks), and GTR + I + G + F for the other data sets (Table 2). The Markov chain Monte Carlo (MCMC) algorithm was run for 2, 000, 000 generations. One tree was sampled every 1000 generations. The first 25% of generations were discarded as burn-in. To construct a consensus tree, we used trees that remained after reaching a stagnant state, i.e., when the average standard deviation of the splitting frequency was < 0.01. Reconstructed trees were visualized in FigTree v.1.4.2 (Rambaut, 2012).
3. Results 3.1. Features of the plastomesAll newly sequenced Salvia and Melissa plastomes possess the typical quadripartite structure of angiosperms, with two single-copy regions (LSC and SSC) and a pair of IR regions (IRa and IRb) (Fig. 1). The length of the plastomes in Salvia ranged from 150, 829 bp (S. yunnanensis) to 154, 984 bp (Salvia castanea Diels f. castanea). The LSC regions ranged from 82, 078 bp in Salvia tiliifolia Vahl to 83, 879 bp in S. sp.; the SSC regions ranged from 17, 500 bp in S. officinalis to 19, 031 bp in Salvia mekongensis Stib.; and the IR regions ranged from 24, 978 bp in S. mekongensis to 27, 265 bp in S. castanea f. castanea. The GC content ranged from 37.9 to 38.1%, with the lowest GC content observed in Salvia subpalmatinervis Stib. and S. mekongensis (37.9%) and the highest GC content observed in S. yunnanensis, S. rosmarinus 1 and S. castanea f. castanea (38.1%). The plastomes of the two Melissa species were 151, 885 bp and 151, 985 bp, and the GC content was 38.0% and 38.1%, respectively (Table 3). All plastomes contained the same 114 unique genes, including 80 protein-coding genes, 30 tRNA genes and 4 rRNA genes (Fig. 1, Table 3).
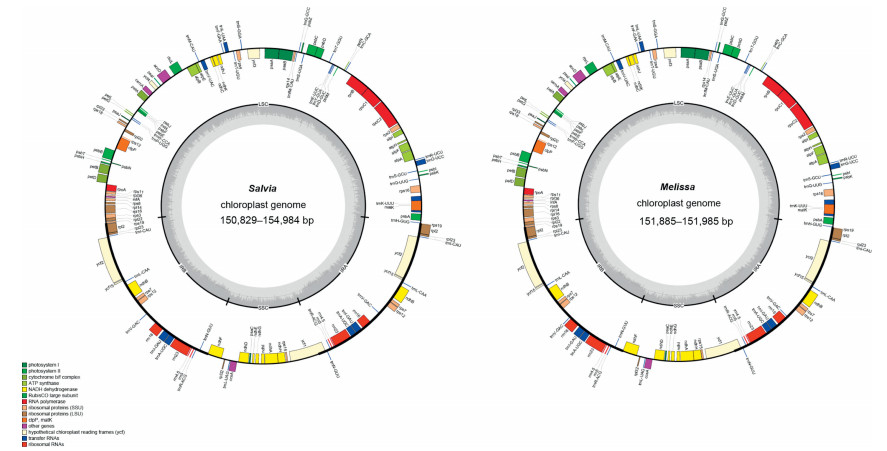
|
| Fig. 1 Plastome gene maps for Salvia and Melissa species. Genes on inside of the map are transcribed in a clockwise direction, whereas genes on the outside of the map are transcribed in a counterclockwise direction. Color coding indicates genes of different functional groups. |
| Taxon | Total length (bp) | LSC (bp) | SSC (bp) | IR (bp) | GC content (%) | Genes (unique) | CDS (unique) | tRNA (unique) | rRNA (unique) |
| Salvia brachyloma | 151, 329 | 82, 567 | 17, 634 | 25, 564 | 38 | 114 | 80 | 30 | 4 |
| S. bulleyana | 151, 547 | 82, 853 | 17, 596 | 25, 549 | 38 | 114 | 80 | 30 | 4 |
| S. castanea f. castanea | 154, 984 | 82, 870 | 17, 584 | 27, 265 | 38.1 | 114 | 80 | 30 | 4 |
| S. castanea f. tomentosa | 151, 589 | 82, 896 | 17, 595 | 25, 549 | 38 | 114 | 80 | 30 | 4 |
| S. cavaleriei | 151, 432 | 82, 638 | 17, 514 | 25, 640 | 38 | 114 | 80 | 30 | 4 |
| S. chanryoenica | 151, 689 | 82, 903 | 17, 634 | 25, 576 | 38 | 114 | 80 | 30 | 4 |
| S. cyclostegia | 151, 644 | 82, 819 | 17, 623 | 25, 601 | 38 | 114 | 80 | 30 | 4 |
| S. flava | 151, 489 | 82, 793 | 17, 608 | 25, 544 | 38 | 114 | 80 | 30 | 4 |
| S. mairei | 151, 629 | 82, 831 | 17, 606 | 25, 596 | 38 | 114 | 80 | 30 | 4 |
| S. mekongensis | 151, 746 | 82, 759 | 19, 031 | 24, 978 | 37.9 | 114 | 80 | 30 | 4 |
| S. miltiorrhiza | 151, 328 | 82, 695 | 17, 555 | 25, 539 | 38 | 114 | 80 | 30 | 4 |
| S. officinalis | 151, 089 | 82, 407 | 17, 500 | 25, 591 | 38 | 114 | 80 | 30 | 4 |
| S. plectranthoides | 151, 416 | 82, 682 | 17, 554 | 25, 590 | 38 | 114 | 80 | 30 | 4 |
| S. przewalskii | 151, 316 | 82, 729 | 17, 605 | 25, 491 | 38 | 114 | 80 | 30 | 4 |
| S. rosmarinus 1 | 151, 695 | 82, 753 | 17, 628 | 25, 657 | 38.1 | 114 | 80 | 30 | 4 |
| S. rosmarinus 2 | 152, 462 | 83, 355 | 17, 965 | 25, 571 | 38 | 114 | 80 | 30 | 4 |
| S. sp. | 152, 658 | 83, 879 | 17, 621 | 25, 579 | 38 | 114 | 80 | 30 | 4 |
| S. subpalmatinervis | 151, 385 | 82, 728 | 17, 605 | 25, 526 | 37.9 | 114 | 80 | 30 | 4 |
| S. tiliifolia | 150, 891 | 82, 078 | 17, 533 | 25, 640 | 38 | 114 | 80 | 30 | 4 |
| S. umbratica | 151, 606 | 82, 728 | 17, 684 | 25, 597 | 38 | 114 | 80 | 30 | 4 |
| S. yunnanensis | 150, 829 | 82, 123 | 17, 572 | 25, 567 | 38.1 | 114 | 80 | 30 | 4 |
| Melissa axillaris | 151, 885 | 83, 000 | 17, 635 | 25, 625 | 38.1 | 114 | 80 | 30 | 4 |
| M. yunnanensis | 151, 985 | 83, 032 | 17, 641 | 25, 656 | 38 | 114 | 80 | 30 | 4 |
In all plastomes, the coding region of the rps19 gene is located at the LSC/IRb boundary, which is stable at 42–43 bp, resulting in a pseudogene fragment (Ψrps19) at the LSC/IRa border. In all plastomes except Salvia mekongensis and S. rosmarinus 2, the ndhF gene spanned the IRb/SSC boundary and ranged from 32 to 64 bp. In all plastomes except S. rosmarinus 1, the coding region of the ycf1 gene spans the IRa/SSC region with a length of 402–1167 bp, which results in a pseudogene (Ψycf1) at the IRa/SSC boundary (Fig. 2). In all plastomes, sequences were more conserved in the IR regions than in the LSC and SSC regions; furthermore, sequence variation in non-coding regions was more divergent than in coding regions. Plastome sequences varied most in the intergenic region (e.g., rps16–trnQ–UUG, atpH–atpI, petN–psbM, psaA–ycf3, accD–psaI, ycf4–cemA, petA–psbJ, rps12–trnV–GAC, rpl32–trnL) (Fig. 3).
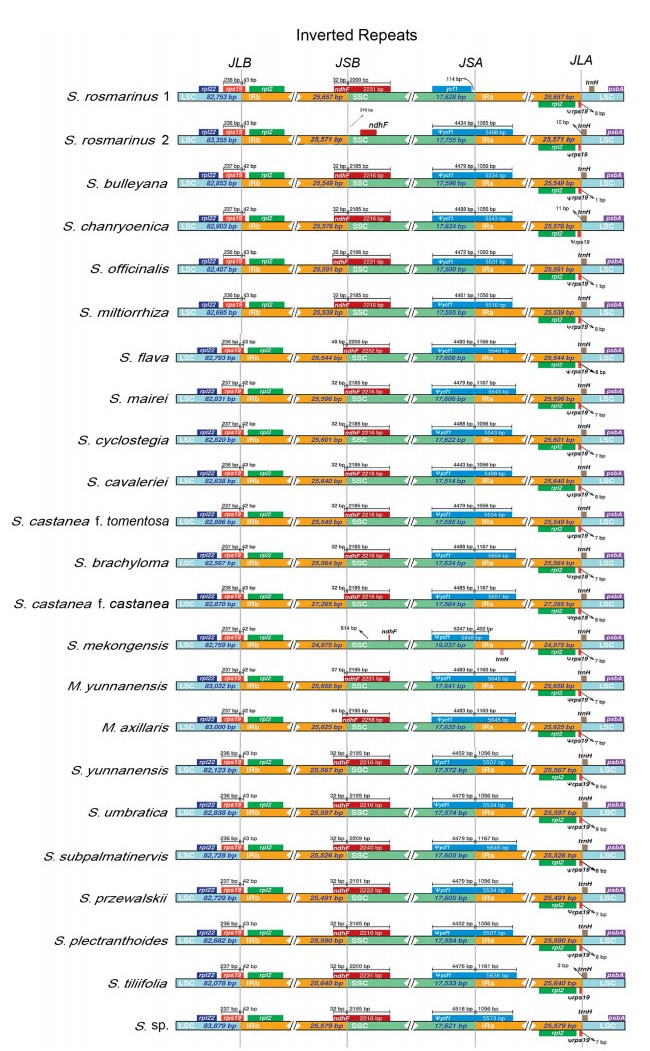
|
| Fig. 2 Comparison of the junctions between the LSC, SSC and IR regions among Salvia and outgroup samples. |
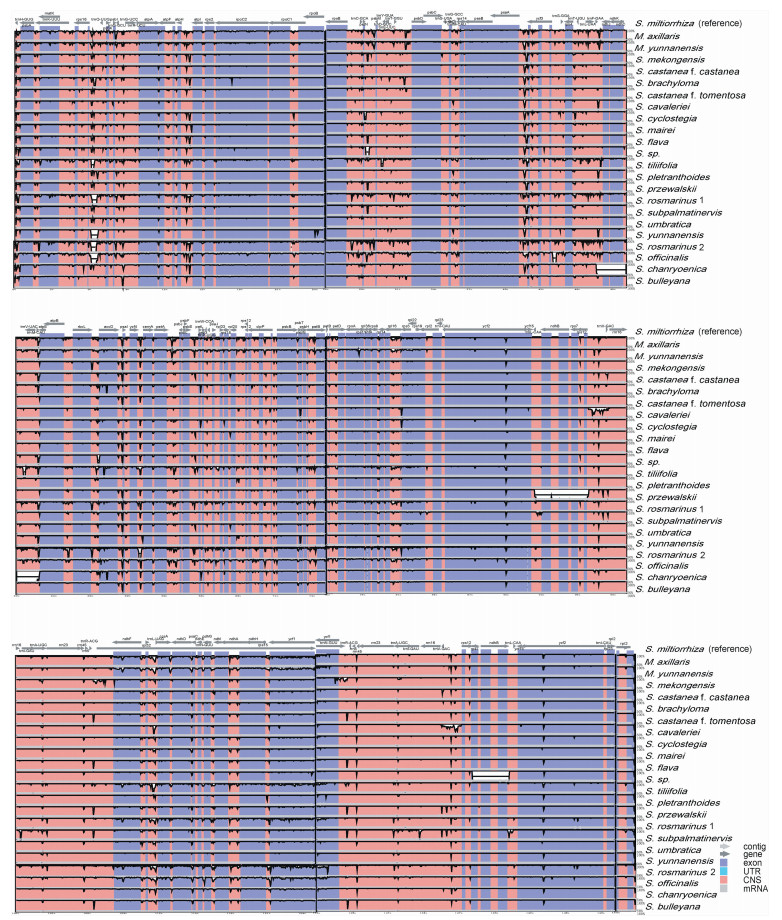
|
| Fig. 3 Sequence alignment of plastomes of Salvia and outgroup samples compared in this study using mVISTA. S. miltiorrhiza was the reference genome. The vertical scale shows the percentage of identity, ranging from 50 to 100%. |
The average nucleotide diversity (Pi) of these plastomes was 0.009383 (Fig. 4). Pi values were mostly greater than 0.008 in the LSC and SSC regions. Moreover, two regions (ycf1 and rpl32–trnL–UAG) with relatively high variability (> 0.03) were located in SSC regions. The highest divergence level in SSC regions was 0.04112 (ycf1) (Fig. 4). In the LSC region, one gene (rpl16) and one intergenic region (trnL–UUU–rps16) had a relatively high variability. The IR regions also had one highly variable locus, rps12–trnV–GAC.
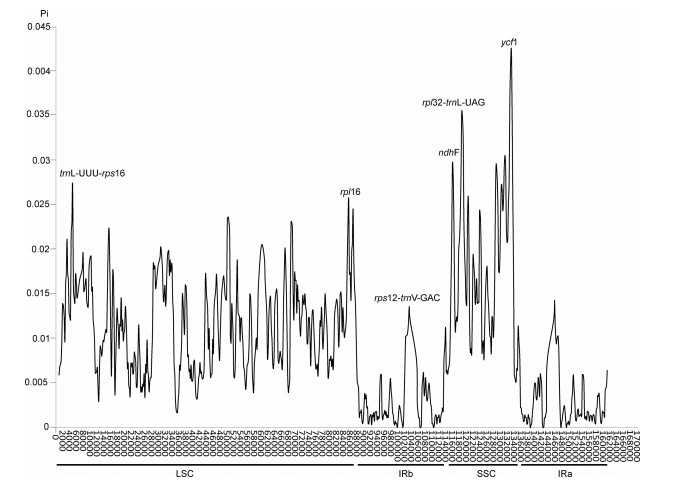
|
| Fig. 4 Sliding window analysis of 23 Lamiaceae plastomes. X-axis: position of the midpoint of a window. Y-axis: nucleotide diversity of each window. Pi of 23 Lamiaceae plastomes. Mutational hotspots and highly divergent loci are marked. |
Ten matrices from 23 complete plastomes were used for phylogenetic analyses (Table 2). The complete plastid DNA sequences only containing one IR (Plastomes) data set was the longest alignment with a length of 137, 226 bp and the shortest alignment was the IR region with Gblocks (IR (Gblocks)) with a length of 24, 540 bp. The constant sites from large to small were plastomes data set with 113, 108 bp to IR (Gblocks) dataset with 23, 993 bp. The parsimony-informative varied from the plastomes dataset with 3644 bp to IR/IR (Gblocks) dataset with 119 bp.
ML, MP and BI analyses resulted in a very similar phylogenetic relationship. The ML topology was therefore selected for discussion, and ML bootstrap (MLBS), MP bootstrap (MPBS), and posterior probabilities (PP) values are given above branches (Fig. 5, Fig. 6, Fig. 7). In this study, we defined well supported as MLBS ≥85%, MPBS ≥85%, and PP ≥ 0.99; moderately supported as 70% < MLBS ≤85%, 70% < MPBS ≤85%, 0.90 < PP ≤ 0.99; and weakly-supported as MLBS < 70%, MPBS < 70%, and PP < 0.90.
|
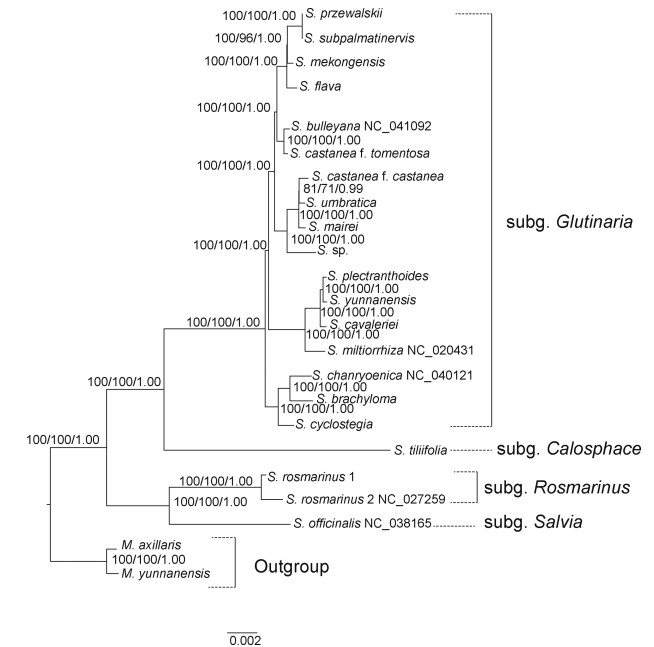
| Fig. 5 Phylogenetic relationships of Salvia based on a data set of sequences from the large single-copy and small single-copy regions with rapidly evolving sites removed (SCR (Gblocks)). Melissa axillaris and M. yunnanensis serve as the outgroup. ML bootstrap (MLBS) values followed by MP bootstrap (MPBS) values and posterior probabilities (PP) values are given above branches. Numbers above the lines indicate MLBS, MPBS and PP values of each clade ≥50%. MLBS, MPBS and PP values < 50% are shown by '-'. |
|
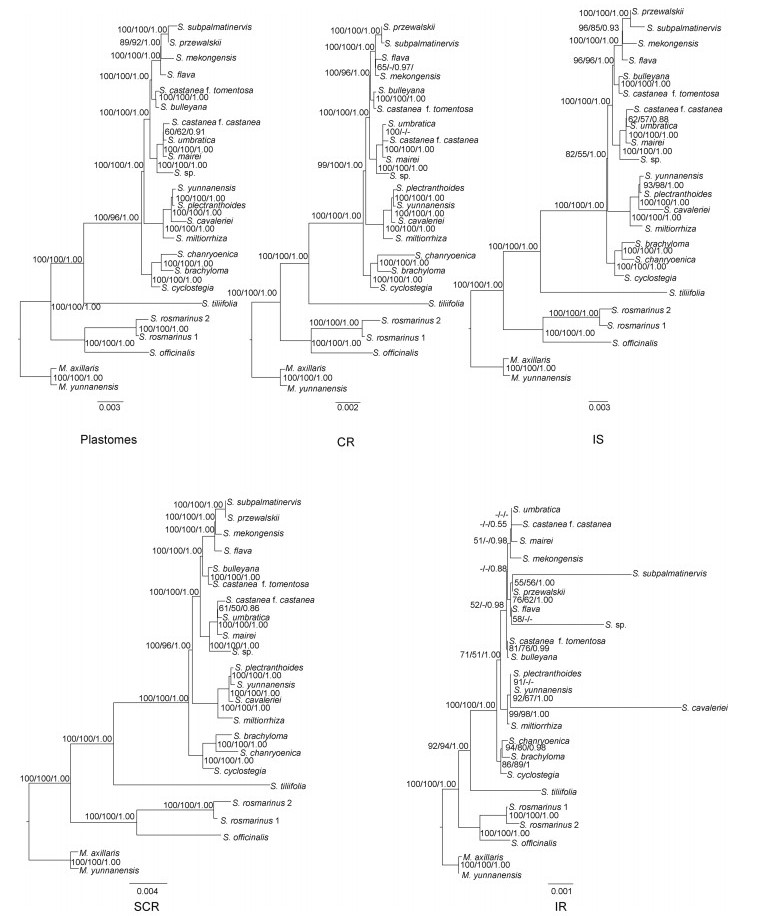
| Fig. 6 Phylogenetic trees of Salvia inferred from analysis of matrix data for 21 plastomes. MLBS, MPBS and PP values are given above branches. Numbers above the lines indicate MLBS, MPBS and PP values of each clade ≥50%. MLBS, MPBS and PP values < 50% are showed by '-'. |

|
| Fig. 7 Phylogenetic trees of Salvia as inferred from analysis of plastome data in which rapidly evolving sites were removed (Gblock). MLBS, MPBS and PP values are given above branches. Numbers above the lines indicate MLBS, MPBS and PP values of each clade ≥50%. MLBS, MPBS and PP values < 50% are showed by '-'. |
In all matrices, Salvia formed a monophyly with strong support (Fig. 6, Fig. 7, MLBS, MPBS, PP = 100%, 100%, 1.00, respectively). Three clades were recovered (Fig. 5). The first clade, which is strongly supported (100%, 100%, 1.00), consists of two subgenera, S. subg. Rosmarinus represented by S. rosmarinus and S. subg. Salvia represented by S. officinalis. The second clade is the Salvia subg. Calosphace represented by Salvia tillifolia. The third clade is the subgenus Glutinaria with strongly supported values (100%, 100%, 1.00), consisting of 16 species (Fig. 5, Fig. 6, Fig. 7). The topologies of ten phylogenetic trees obtained by different matrices are highly similar, with only the support values varying in different trees (Fig. 6, Fig. 7). The phylogenetic tree based on the matrix of LSC and SSC region with Gblocks (SCR(Gblocks)) had the highest support values. All relationships were well supported (100%, 96–100%, 1.00) with the exception of that between Salvia umbratica Hance and S. castanea f. castanea, which was moderately supported (Fig. 7, 81%, 71%, 0.99). The poorest resolved phylogenetic trees were constructed using IR and IR (Gblocks); some branches had weak-support, and only five relationships were strongly supported (100%, 100%, 1.00) (Fig. 6, Fig. 7).
4. Discussion 4.1. Genome organizationAll Salvia plastomes examined here possess the highly conserved quadripartite structure typical of most angiosperms. Salvia plastomes shared 114 unique genes that include 80 protein-coding genes, 30 tRNAs, and 4 rRNAs (Lukas and Novak, 2013), which is consistent with the plastomes of two Melissa (Lamiaceae, Nepetoideae) species and Lamiaceae plastomes such as Lavandula angustifolia Mill. (Lamiaceae, Nepetoideae) (Ma, 2018). Salvia plastome gene order is consistent with that of species throughout Lamiaceae (He et al., 2016; Ma, 2018). Moreover, the 21 Salvia plastomes had similar GC content. Furthermore, the GC content of Saliva was to those of other genus within the Lamiaceae family, even as well as to that of Boea hygrometrica (Bunge) R. Br. in Gesneriaceae family from the same order (Lamiales) (Zhang et al., 2012).
In angiosperms, gene loss-and-gain events and structural rearrangement of plastomes have been found in some species or genera (Wicke et al., 2011), but these events have not been observed in Salvia species. These results suggest that the plastome structure and content of Salvia are largely conservative, even in the order Lamiales.
IR contraction-expansion is a pervasive and primary cause of plastome length variation (Raubeson et al., 2007; Wang et al., 2008). The IR/LSC junctions of the 21 Salvia plastomes were highly conserved (Fig. 2). In all Salvia plastomes, the Ψrps19 and trnH genes were present at the IRs/LSC boundaries. The IRb/SSC junctions of 19 Salvia plastomes were conserved, and the ndhF gene spanned the IRb/SSC junctions. IR contraction was observed in two species (S. rosmarinus 2 and S. mekongensis). The IRa/SSC junctions of the 20 Salvia plastomes were conserved and the ycf1 gene spanned the IRa/SSC border, IR contraction was only observed found in S. rosmarinus 1. The SSC/IR boundaries of the two S. rosmarinus accessions differed, which may be due to intraspecies variation. The largest difference in Salvia plastome size was 4155 bp (S. yunnanensis versus S. castanea f. castanea); in the IR region the largest difference was 2287 bp (S. mekongensis versus S. castanea f. castanea); in the SSC region, the largest difference was 1531 bp (S. officinalis versus S. mekongensis); and in the LSC region, the largest difference was 1801 bp (S. tiliifolia versus S. sp.). These differences in Salvia plastid genome lengths can be largely explained by differences in IR length, which was the region of greatest variation. Salvia plastome sequence variation was highly consistent (Fig. 3); IR regions were mostly conserved and non-coding regions were more highly divergent than coding regions. These patterns suggest that the content and structure of Salvia plastomes are highly conserved. The Pi values of these sites (ycf1 and rpl32–trnL–UAG) exceed 0.03, with one Pi value, ycf1, higher than 0.04. We believe that these sites with large variation can be used to identify species in Salvia.
4.2. Phylogenetic implicationsIn this study, we used ten data matrices to generate high resolution phylogenetic trees to evaluate the effectiveness of different plastome sequences in phylogenetic inference of Salvia, and in particular the Salvia subg. Glutinaria. All phylogenetic trees have highly similar topologies and are generally well supported (Fig. 5, Fig. 6, Fig. 7). Recently, East Asian Salvia have been treated as a subgenus, S. subg. Glutinaria (Hu et al., 2018). Our results comfirm that S. subg. Glutinaria is a monophyletic group with strong support in all trees (Fig. 5, Fig. 6, Fig. 7), corroborating previous work based on two nuclear ribosomal spacers and four plastid markers (Hu et al., 2018). Previous studies have been unable to determine the interspecific relationships between species within the subg. Glutinaria (e.g., S. cavaleriei, S. miltiorrhiza and S. yunnanensis) (Hu et al., 2018). In our study, S. cavaleriei, S. miltiorrhiza and S. yunnanensis form a strongly supported clade with Salvia plectranthoides, in which S. miltiorrhiza occupies the basal position, followed by S. cavaleriei, which is sister to S. yunnanensis and S. plectranthoides in all trees. The phylogenetic trees generated in our study are more highly resolved than those generated by nuclear genes in previous studies and differ in several ways (Hu et al., 2018). 1) Salvia cyclostegia is not in sect. Eurysphace (which includes S. przewalskii, S. subpalmatinervis, S. bulleyana, S. castanea, S. mairei, and Salvia flava). 2) S. cavaleriei is embedded in sect. Drymosphace (which includes S. miltiorrhiza, S. yunnanensis, and S. plectranthoides), and 3) S. umbratica is embedded in sect. Eurysphace. Many factors may contribute to these discrepancies, including incomplete lineage sorting, chloroplast capture, and hybridization (Albaladejo et al., 2005; Deng et al., 2015; Xiang et al., 2013). S. castanea Diels f. tomentosa and S. castanea f. castanea do not form a monophyletic group, which is consistent with previous phylogenies based on nuclear ribosomal spacers (Hu et al., 2018). This implies that these two sub-species underwent cryptic speciation rather than hybridization, as the two forms are morphologically similar, but genetically distinct. Previous molecular phylogenetic studies have used both nuclear genome fragments and plastid fragments; however, clades were not highly supported and relationships were not well resolved (Drew et al., 2017; Hu et al., 2018; Will and Classen-Bockhoff, 2017). The main reason for low support of previous studies is the lack of a sufficient number of informative sites to construct phylogenetic tree. The use of plastid phylogenomics in the present study has provided better resolution of the phylogenetic tree of Salvia subg. Glutinaria.
Phylogenetic analysis using complete plastome sequences contained over 100 times more simplified informative sites than those of previous analysis based on DNA fragments. Consequently, each node of our phylogenetic trees is highly supported. Specifically, four matrices have high support (Fig. 6, Fig. 7) (Plastomes, SCR, Plastomes (Gblocks), and SCR (Gblocks)), and only one or two nodes do not have strongly supported values (100%, 100%, 1.00). The other six matrices all have more than two nodes weakly supported. These analyses indicate that plastome sequences are conducive to resolving phylogenetic relationships of Salvia subg. Glutinaria. Saturation of sequence mutations can impact support values of phylogenetic trees (Knight and Mindell, 1993; Simon et al., 1994). From the constructed phylogenetic trees, we found that resolving phylogenetic relationships is not a matter of having the most variable sites possible. Some saturated variation will affect the resolution of systematic relationships, a phenomenon that now seems to be evident in Salvia. For example, although the plastome matrix had the greatest number of informational sites (3644 bp), two relationships are still only moderately supported. Phylogenetic trees based on the SCR matrix, which had a lower number of informational sites (3505 bp), have higher support values, and only the relationship between S. castanea f. castanea and S. umbratica was weakly-supported (Fig. 6, 61%, 50%, 0.86). Gblocks reduced the number of informative sites in the SCR (Gblocks) data set (3425 bp); however, the clade that contains S. castanea f. castanea and S. umbratica became moderately supported (Fig. 7, 81%, 71%, 0.99). Conversely, there can be too few informative sites. Most clades based on IR and IR (Gblocks) (119 bp) were weakly supported because there were insufficient numbers of informative sites (Table 2). Previous studies have suggested that the selection of suitable information sites is important to phylogenetic reconstruction. For example, in Cymbidium Sw., using introns and interval sequences of plastomes can effectively enhance the resolution of the tree (Yang et al., 2013a). Here the phylogenetic tree constructed with the SCR (Gblocks) matrix resolved relationships within the Salvia subg. Glutinaria very well (Fig. 6, Fig. 7). Therefore, our findings show that using sequences with suitable mutations and informative sites is helpful for resolving phylogenetic relationships. In fact, because saturation of sequence mutations may affect phylogenetic reconstruction, deleting saturation mutation information appropriately is more conducive for phylogenetic reconstruction.
Although all phylogenetic relationships cannot be resolved by using complete plastomes (Petersen et al., 2011; Richard et al., 2009), our results suggest that plastome-wide analysis will provide resolution for some disputed relationships. Further research is needed that combines nuclear genes and more plastomes to elucidate the phylogeny and evolution of Salvia.
5. ConclusionThe present study has shown that the plastome structure of Salvia is largely conserved. The plastome of 21 Salvia has a typical quadripartite structure with 114 coding genes. The length of plastomes in Salvia is 150, 829–154, 984 bp, GC content is 37.9–38.1%, and the variation of the non-coding regions is higher than coding regions. These characteristics are similar to those of the plastomes of other Lamiaceae species. We found that using complete plastomes can better resolve the phylogenetic relationships of Salvia subg. Glutinaria. In addition, our results show that using a plastome data set that includes the large single-copy and small single-copy regions with rapidly evolving sites removed is the most suitable approach to reconstructing the phylogeny of Salvia. Our demonstration that comparative plastomic analysis provides insight into the phylogeny of Salvia, the largest genus of Lamiaceae, indicating that using suitable mutations with informative sites are helpful in resolving phylogenetic problems.
Author contributionsStudy conception: HW, H-TL, D-ZL; acquisition of molecular data: HW, H-TL; plastomic and phylogenetic analysis: HW, H-TL, P-FM; drafting of manuscript and pictures: HW, H-TL, P-FM, G-XH, D-ZL; critical revisions: HW, P-FM, D-ZL. All authors have read, commented and approved the final manuscript.
Declaration of Competing InterestThe authors declared that they have no conflict of interest.
AcknowledgementsWe are grateful to the Germplasm Bank of Wild Species at the Kunming Institute of Botany (KIB) and Molecular Biology Experiment Center, Germplasm Bank of Wild Species for facilitating this study. This research was supported by the Large-scale Scientific Facilities of the Chinese Academy of Sciences (Grant No: 2017–LSFGBOWS–02). We thank Dr. Chun-Lei Xiang for reviewing the first draft for us.
A Frazer K., Pachter L., Poliakov A., et al, 2004. VISTA: computational tools for comparative genomics. Nucleic Acids Res, 32: W273-W279. DOI:10.1093/nar/gkh458 |
Albaladejo R.G., Aguilar J.F., Aparicio A., et al, 2005. Contrasting nuclear-plastidial phylogenetic patterns in the recently diverged Iberian Phlomis crinita and P. lychnitis lineages (Lamiaceae). Taxon, 54: 987-998. DOI:10.2307/25065483 |
Ali N.M., Yeap S.K., Ho W.Y., et al, 2012. The promising future of chia, Salvia hispanica L.. J. Biomed. Biotechnol, 2012: 1-9. |
Amiryousefi A., Hyvönen J., Poczai P., 2018. IRscope: an online program to visualize the junction sites of chloroplast genomes. Bioinformatics, 34: 3030-3031. DOI:10.1093/bioinformatics/bty220 |
Castresana J., 2000. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol. Biol. Evol, 17: 540-552. DOI:10.1093/oxfordjournals.molbev.a026334 |
Chen S.L., Song J.Y., Yao H.I., et al, 2009. Identification strategies and key techniques of DNA barcoding for medicinal plants. Chin. J. Nat. Med, 7: 322-327. |
ClaßeneBockhoff R., Wester P., Tweraser E., 2008. The staminal lever mechanism in Salvia L. (Lamiaceae)-a review. Plant Biol, 5: 33-41. |
Deng T., Nie Z.-L., Drew B.T., et al, 2015. Does the Arcto-Tertiary biogeographic hypothesis explain the disjunct distribution of northern hemisphere herbaceous plants? The case of Meehania (Lamiaceae). PloS One, 10: e0117171. DOI:10.1371/journal.pone.0117171 |
Doyle J.J., 1987. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem. Bull, 19: 11-15. |
Drew B., González-Gallegos J.G., Xiang C.-L., et al, 2017. Salvia united: the greatest good for the greatest number. Taxon, 66: 133-145. DOI:10.12705/661.7 |
Fragoso-Martínez I., Salazar G.A., Martínez-Gordillo M., et al, 2017. A pilot study applying the plant anchored hybrid enrichment method to new world sages(Salvia subgenus Calosphace; Lamiaceae). Mol. Phylogenet. Evol, 117: 124-134. DOI:10.1016/j.ympev.2017.02.006 |
Fragoso-Martínez I., Martínez-Gordillo M. Salazar G.A., et al, 2018. Phylogeny of the Neotropical sages (Salvia subg. Calosphace; Lamiaceae) and insights into pollinator and area shifts. Plant Syst. Evol, 304: 1-13. DOI:10.1007/s00606-017-1439-2 |
Ge X.X., Chen H.W., Wang H.L., et al, 2014. De novo assembly and annotation of Salvia splendens transcriptome using the Illumina platform. PloS One, 9: e87693. DOI:10.1371/journal.pone.0087693 |
He Y., Xiao H.Y., Deng C., Xiong L., et al, 2016. The complete chloroplast genome sequences of the medicinal plant Pogostemon cablin. Int. J. Mol. Sci, 17: 820. DOI:10.3390/ijms17060820 |
He Y.-H., Han L.M., Liu Y.P., et al, 2017. Complete sequence analysis of chloroplast genome of Salvia japonica. Bull. Bot. Res, 37: 572-578. |
Hu G.X., Takano A., Drew B.T., Liu E.D., et al, 2018. Phylogeny and staminal evolution of Salvia (Lamiaceae, Nepetoideae) in East Asia. Ann. Bot, 122: 649-668. DOI:10.1093/aob/mcy104 |
Jenks A.A., Walker J.B., Seung-Chul K., 2012. Phylogeny of new world Salvia subgenus Calosphace (Lamiaceae) based on cpDNA (psbA-trnH) and nrDNA (ITS)sequence data. J. Plant Res, 126: 483-496. DOI:10.1007/s10265-012-0543-1 |
Jin J.-J., Yu W.-B., Yang J.-B., et al, 2018. GetOrganelle: a fast and versatile toolkit for accurate de novo assembly of organelle genomes. BioRxiv: 256479. |
Kalyaanamoorthy S., Minh B.Q., Wong T.K., et al, 2017. ModelFinder: fast model selection for accurate phylogenetic estimates. Nat. Methods, 14: 587-589. DOI:10.1038/nmeth.4285 |
Katoh K., Kuma K.I., Miyata T., et al, 2005. Improvement in the accuracy of multiple sequence alignment program MAFFT. Genome Inf, 16: 22-33. |
Knight A., Mindell D., 1993. Substitution bias, weighting of dna sequence evolution, and the phylogenetic position of fea's viper. Syst. Biol, 42: 18-31. DOI:10.1093/sysbio/42.1.18 |
Kriebel R., Drew B.T., Drummond C., et al, 2019. Tracking temporal shifts in area, biomes, and pollinators in the radiation of Salvia (sages) across continents: leveraging anchored hybrid enrichment and targeted sequence data. Am. J. Bot, 106: 573-597. DOI:10.1002/ajb2.1268 |
Li Q.Q., Li M.H., Yuan Q.J., et al, 2013. Phylogenetic relationships of Salvia (Lamiaceae) in China: evidence from DNA sequence datasets. J. Syst. Evol, 51: 184-195. DOI:10.1111/j.1759-6831.2012.00232.x |
Librado P., Rozas J., 2009. DnaSP v5:a software for comprehensive analysis of DNA polymorphism data. Bioinformatics, 25: 1451-1452. DOI:10.1093/bioinformatics/btp187 |
Lin C.P., Wu C.S., Huang Y.Y., et al, 2012. The complete chloroplast genome of Ginkgo biloba reveals the mechanism of inverted repeat contraction. Genome Biol. Evol, 4: 374-381. DOI:10.1093/gbe/evs021 |
Lindqvist C., Albert V.A., 2002. Origin of the Hawaiian endemic mints within north American Stachys (Lamiaceae). Am. J. Bot, 89: 1709-1724. DOI:10.3732/ajb.89.10.1709 |
Lowe T.M., Chan P.P., 2016. tRNAscan-SE on-line: integrating search and context for analysis of transfer RNA genes. Nucleic Acids Res, 44: W54-W57. DOI:10.1093/nar/gkw413 |
Lukas B., Novak J., 2013. The complete chloroplast genome of Origanum vulgare L. (Lamiaceae). Gene, 528: 163-169. DOI:10.1016/j.gene.2013.07.026 |
Ma L., 2018. The complete chloroplast genome sequence of the fragrant plant Lavandula angustifolia (Lamiaceae). Mitochondrial DNA Part B, 3: 135-136. DOI:10.1080/23802359.2018.1431067 |
Patel R.K., Jain M., 2012. NGS QC Toolkit: a toolkit for quality control of next generation sequencing data. PloS One, 7: e30619. DOI:10.1371/journal.pone.0030619 |
Petersen G., Aagesen L., Seberg O., et al, 2011. When is enough, enough in phylogenetics? A case in point from Hordeum (Poaceae). Cladistics-Int. J. Willi Hennig Soc, 27: 428-446. DOI:10.1111/j.1096-0031.2011.00347.x |
Qian Z., Liang X., Hou A., et al, 2002. Medicinal resources of Salvia yunnanensis. J. Chin. Med. Mater, 25: 628-629. |
Qian J., Song J., Gao H., et al, 2013. The complete chloroplast genome sequence of the medicinal plant Salvia miltiorrhiza. PloS One, 8: e57607. DOI:10.1371/journal.pone.0057607 |
Qu X.J., Moore M.J., Li D.Z., et al, 2019. PGA: a software package for rapid, accurate, and flexible batch annotation of plastomes. Plant Methods, 15: 50. DOI:10.1186/s13007-019-0435-7 |
Rambaut, A., 2012. FigTree V1.4.2. http://tree.bio.ed.ac.uk/software/figtree/.
|
Raubeson L.A., Peery R., Chumley T.W., et al, 2007. Comparative chloroplast genomics: analyses including new sequences from the angiosperms Nuphar advena and Ranunculus macranthus. BMC Genom, 8: 174. DOI:10.1186/1471-2164-8-174 |
Richard C., Matthew P., Aaron L., 2009. Increasing phylogenetic resolution at low taxonomic levels using massively parallel sequencing of chloroplast genomes. BMC Biol, 7: 84. DOI:10.1186/1741-7007-7-84 |
Ronquist F., Teslenko M., Der Mark P.V., et al, 2012. MrBayes 3. 2:efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol, 61: 539-542. DOI:10.1093/sysbio/sys029 |
Simon C., Frati F., Beckenbach A., et al, 1994. Evolution, weighting, and phylogenetic utility of mitochondrial gene sequences and a compilation of conserved polymerase chain reaction primers. Ann. Entomol. Soc. Am, 87: 651-701. DOI:10.1093/aesa/87.6.651 |
Stamatakis A., Ott M., Ludwig T., 2005. RAxML-OMP: an efficient program for phylogenetic inference on SMPs. Lect. Notes Comput. Sci, 3606: 288-302. |
Swofford, D.L., 2002. PAUP*. Phylogenetic Analysis Using Parsimony (*and Other Methods). Version 4. Sinauer Associates, Sunderland, Massachusetts.
|
Takano A., Okada H., 2011. Phylogenetic relationships among subgenera, species, and varieties of Japanese Salvia L. (Lamiaceae). J. Plant Res, 124: 245-252. DOI:10.1007/s10265-010-0367-9 |
Tian X., Li D.Z., 2002. Application of DNA sequences in plant phylogenetic study. Acta Bot. Yunnanica, 24: 170-184. |
Walker J.B., Sytsma K.J., 2006. Staminal evolution in the genus Salvia (Lamiaceae): molecular phylogenetic evidence for multiple origins of the staminal lever. Ann.Bot, 100: 375-391. |
Walker J.B., Sytsma K.J., Treutlein J., et al, 2004. Salvia (Lamiaceae) is not monophyletic: implications for the systematics, radiation, and ecological specializations of Salvia and tribe Mentheae. Am. J. Bot, 91: 1115-1125. DOI:10.3732/ajb.91.7.1115 |
Wang B.Q., 2010. Salvia miltiorrhiza: chemical and pharmacological review of a medicinal plant. J. Med. Plants Res, 425: 2813-2820. |
Wang R.J., Cheng C.L., Chang C.C., et al, 2008. Dynamics and evolution of the inverted repeatelarge single copy junctions in the chloroplast genomes of monocots. BMC Evol. Biol, 8: 36. DOI:10.1186/1471-2148-8-36 |
Wang Y., Zhan D.F., Jia X., et al, 2016. Complete chloroplast genome sequence of aquilaria sinensis (lour.) Gilg and evolution analysis within the malvales order. Front. Plant Sci, 7: 280. |
Wei Y.K., Wang Q., Huang Y.B., 2015. Species diversity and distribution of Salvia(Lamiaceae). Biodivers. Sci, 23: 3-10. DOI:10.17520/biods.2014070 |
Wick R.R., Schultz M.B., Zobel J., et al, 2015. Bandage: interactive visualization of de novo genome assemblies. Bioinformatics, 31: 3350-3352. DOI:10.1093/bioinformatics/btv383 |
Wicke S., Schneeweiss G.M., Depamphilis C.W., et al, 2011. The evolution of the plastid chromosome in land plants: gene content, gene order, gene function. Plant Mol. Biol, 76: 273-297. DOI:10.1007/s11103-011-9762-4 |
Will M., Classen-Bockhoff R., 2017. Time to split Salvia s. l. (Lamiaceae)-new insights from old world Salvia phylogeny. Mol. Phylogenet. Evol, 109: 33-58. DOI:10.1016/j.ympev.2016.12.041 |
Will M., Claßen-Bockhoff R., 2014. Why Africa matters: evolution of old world Salvia (Lamiaceae) in Africa. Ann. Bot, 114: 61-83. DOI:10.1093/aob/mcu081 |
Wyman S.K., Jansen R.K., Boore J.L., 2004. Automatic annotation of organellar genomes with DOGMA. Bioinformatics, 20: 3252-3255. DOI:10.1093/bioinformatics/bth352 |
Xiang C.-L., Zhang Q., Scheen A.-C., et al, 2013. Molecular phylogenetics of Chelonopsis (Lamiaceae: gomphostemmateae) as inferred from nuclear and plastid DNA and morphology. Taxon, 62: 375-386. DOI:10.12705/622.11 |
Yang J.B., Tang M., Li H.T., et al, 2013a. Complete chloroplast genome of the genus Cymbidium: lights into the species identification, phylogenetic implications and population genetic analyses. BMC Evol. Biol, 13: 84. DOI:10.1186/1471-2148-13-84 |
Yang J.B., Yang S.X., Li H.T., et al, 2013b. Comparative chloroplast genomes of Camellia species. PloS One, 8: e73053. DOI:10.1371/journal.pone.0073053 |
Zhang H.J., Li L.N., 1994. Salvianolic acid I: a new depside from Salvia cavaleriei. Planta Med, 60: 70-72. DOI:10.1055/s-2006-959411 |
Zhang T., Fang Y., Wang X., et al, 2012. The complete chloroplast and mitochondrial genome sequences of Boea hygrometrica: insights into the evolution of plant organellar genomes. PloS One: 7. |
Zhang D., Gao F., Jakovlić I., et al, 2020. PhyloSuite: an integrated and scalable desktop platform for streamlined molecular sequence data management and evolutionary phylogenetics studies. Mol. Ecol. Resour, 20: 348-355. DOI:10.1111/1755-0998.13096 |



