b. University of the Chinese Academy of Sciences, Beijing 100049, China;
c. College of Pharmaceutical Science, Yunnan University of Chinese Medicine, Kunming 650500, China
Understanding the origin and evolution of endemic species has long intrigued biogeographers, evolutionary biologists, and molecular ecologists (Gaston, 2000; Lohmann et al., 2013). Accordingly, researchers have increasingly examined the flora of the mountains of Central Asia, including the Himalayas and the Hengduan Mountains, which are unique biodiversity hotspots (Myers et al., 2000; Mittermeier et al., 2011). These mountains surround the Qinghai-Tibet Plateau (QTP), the world's highest and most extensive plateau (Zhang et al., 2002). The ongoing uplift of the plateau, which started 25 million years ago (Ma), has had a major influence on plant diversity in the region (Harrison et al., 1992; Li et al., 1995; Guo et al., 2002; Spicer et al., 2003). Specifically, the QTP uplift has created a large elevational gradient that fragments the habitat of alpine plant species, creating barriers to gene flow that increase divergent evolution and speciation (Filatov et al., 2016). As a result, the uplift of the QTP has created young endemic 'plant cradles' in the Hengduan Mountains, which act as an 'evolutionary front' in China (Lopez-Pujol et al., 2011). However, the origin and evolution of many montane species (e.g., Incarvillea spp.) and the biogeographical patterns (e.g., dispersal routes) between the mountainous hotspots of Central Asia have yet to be fully studied; therefore, understanding the causes and consequences of these patterns remains a challenge (e.g., Nauheimer et al., 2012).
To date, few studies have investigated the connections between the QTP and adjoining biodiversity hotspots in Asia. The similarity of floristic elements and geographic distribution patterns of plants that occurs throughout Asia may reflect evidence of dispersal of montane plants to Southeast Asia from the Himalaya region, as well as Central Asia, or the reverse.
This unusual biogeographical connection may be explained by long-distance dispersal that uses SE Asia as a 'stepping stone' (Liu et al., 2002). The QTP has acted as both a source and a sink for plant taxa that occur in the Himalaya and adjoining regions, the New and Old World, the Middle East, as well Indochina (Hajra and Rao, 1990; Zhu and Roos, 2004; Xu et al., 2014). Surprisingly, research has shown that the 'highlands of Central Asia' (Li et al., 2014) and the 'Beringia' land bridge connecting Eastern Asia with Northern America (Nie et al., 2006; Deng et al., 2018) have acted as corridors for plant genera that originated in the QTP (Xu et al., 2010; Zhang et al., 2014). However, evidence suggests that plant taxa may have originated outside the QTP and diversified after their ancestors arrived to the QTP region (Liu et al., 2002; Yue et al., 2009; Tu et al., 2010). In addition, the QTP might serve as a 'refugium' for Tertiary relict floras (Milne and Abbott, 2002).
Incarvillea Juss. is a temperate and herbaceous montane genus that comprises 16 species distributed throughout the western, southern and eastern fringe of the QTP (Grierson, 1961; Zhao, 1985; Zhang and Santisuk, 1998; Chen et al., 2005). Recent molecular and taxonomic work by Chen et al.(2005, 2006) revealed five major subgenera, including the subgenus Olgaea (Table 1). Of these subgenera, the subgenus Incarvillea is distributed mainly in East Asia (I. sinensis Lam.: from SW to NE China) and Mongolia (I. potaninii Batalin), whereas the remaining four subgenera are distributed at various elevations (500–5500 m a.s.l.) in the three major biodiversity hotspots of Asia, i.e. the Mountains of Central Asia (subgenera Olgaea and Niedzwedzkia), the Himalaya (subgenera Amphicome and Pteroscleris), and Mountains of SW China (subgenus Pteroscleris) (Chen et al., 2005) (Fig. 1 a). Previous research on Incarvillea has focused on molecular diversity and species richness assessments (genus Incarvillea; Chen et al., 2010), population genetic analysis (I. younghusbandii Sprague; Zhu et al., 2009: I. sinensis; Chen et al., 2012: I. arguta (Royle) Royle; Rana et al., 2019), pollen dispensing mechanisms (I. arguta; Han et al., 2008) and reproductive strategies of a pollinator-limited species (I. mairei (H.Lev) Grierson; Ai et al., 2013). However, few studies have examined how, when and where the species of this genus originated and what stochastic characters led to the evolution of the different subgenera.
| Characteristics | Incarvillea subgenus Niedzwedzkia | I. subgenus Amphicome | I. subgenus Incarvillea | I. subgenus Pteroscleris | I. subgenus Olgaea |
| Habit | Suffruticose | Suffruticose | Suffruticose | Herbaceous | Suffruticose |
| Leaf arrangement | Alternate | Alternate | Alternate | Alternate or radical | Opposite |
| Calyx teeth base | Not swollen | Not swollen | Swollen | Not swollen | Not swollen |
| Inflorescence | Racemose | Racemose | Racemose | Racemose | Paniculate |
| Anther texture | Glabrous | Pilose | Glabrous | Glabrous | Glabrous |
| Capsule shape | Ovate | Elongate, linear cylindrical | Cylindrical | Cylindrical | Cylindrical |
| Capsule texture | Subligneous | Fibrous | Coriaceous | Subligneous | Coriaceous |
| Capsule wing | Six wings, longitudinal | No | No | No | No |
| Capsule dehiscence | Septifragal | Loculicidal | Loculicidal | Loculicidal | Loculicidal |
| Seed wing | Opaque wing | Coma at ends | Hyaline wing | Opaque wing | Hyaline wing |
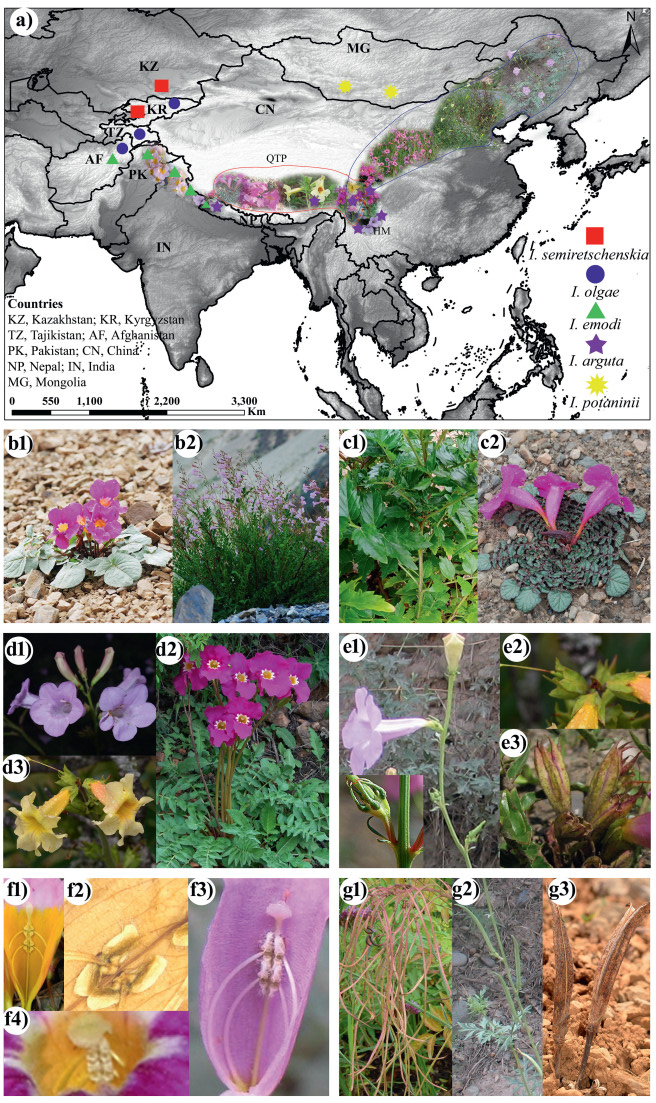
|
| Fig. 1 a) Geographic distribution and pictorial representation of the East Asian subgenus Pteroscleris (red solid line) in the Qinghai-Tibet Plateau (QTP)–Hengduan Mountain (HM) and Incarvillea sinensis (blue solid line), along with (b–g) morphological characters of the montane genus Incarvillea used in character evolution. The morphological following morphological characters are shown: Habitat (b1-c2), Inflorescence (d1–d3), Calyx teeth base (e1–e3), Anther texture (f1–f4), and Capsule shape (g1–g3). Character mapping of the following Incarvillea species is shown: I. arguta (b2, c1, d1, f2, f3, g1), I. compacta (e3, g3), I. emodi (f1), I. lutea (d3, e2), I. mairei var. mairei (b1), I. mairei var. grandiflora (f4), I. sinensis (e1, g2), I. younghusbandii (c2), I. zhongdianensis (d2). Photographs by: Hang Sun: I. compacta (e3); Niu Yang: I. arguta (b2), I. lutea (d3, e2), I. mairei (b1, f4); Shaotian Chen: I. sinensis (e1, g2), I. younghusbandii (c2), I. zhongdianensis (d2); Santosh Kumar Rana: I. arguta (c1, f2, f3, g1), I. emodi (f1); Ya-Zhou Zhang: I. arguta (d1); Qia Wang: I. compacta (g3). |
Molecular phylogenetic analysis of the genus Incarvillea based on ITS and trnL-trnF sequences has provided strong evidence that species within subgenus Pteroscleris might have undergone a recent radiation, perhaps related to the uplift of the Himalaya-Hengduan Mountains (Chen et al., 2005). Thus, members of the genus Incarvillea provide an excellent opportunity to use biogeography to examine species adaptation to recent diversification and rapid radiation. Furthermore, because the five subgenera of Incarvillea clearly differ in their habit, stamens, capsule texture, and seeds (Chen et al., 2004), morphological character evolution may help understand the phylogenetic placement of subgenera.
In this study, we aim to identify the historical biogeography and ancestral characters that have resulted in present-day patterns of Incarvillea diversification. Specifically, we asked when, where and how the genus Incarvillea originated and what conspicuous ancestral characters led to the evolution of the five subgenera of Incarvillea. To answer these questions, we reconstructed a phylogenetic framework of the genus Incarvillea. Our molecular tree incorporated plastid trnL-trnF, nuclear ribosomal Internal Transcribed Spacer (nrITS), and combined sequences from the genus Incarvillea, along with large trnL-trnF sequences from the Bignoniaceae family (except Bignonieae). We then combined information from our time-calibrated tree derived from our molecular data with that of biogeographical considerations, morphological character evolution, and the distribution of representatives of extant Incarvillea species to discuss the events that underlie the evolutionary history of the genus Incarvillea.
2. Material and methods 2.1. Taxon sampling and DNA sequencingWe sampled ca. 81% of extant species of the genus Incarvillea, including representative species from five Incarvillea subgenera. Specifically, we used 18 GenBank accessions representing 13 species of the genus Incarvillea (except Incarvillea altissima Forrest, I. forrestii Fletcher, and I. potaninii). We assume that the generic coverage of the genus Incarvillea represents the overall distribution range (Fig. 1 a), which is likely to lead the reliable reconstruction of the biogeographic history.
New sequences generated in this study include Incarvillea arguta nrITS sequences (MT533886–MT533889) from four individuals representing the entire geographic range of the species (Fig. 1a). Voucher specimens were deposited in the National Herbarium and Plant Laboratories (KATH), Nepal and Kunming Institute of Botany (KUN), China.
Outgroups comprise the tribes or clades within the family Bignoniaceae (except clade Bignonieae) (Grose and Olmstead, 2007; Olmstead et al., 2009; Refulio-Rodriguez and Olmstead, 2014): Tecomeae tribe (9 genera), Catalpeae tribe (3 genera), Tabebuia alliance (9 genera), Plaeotropical clade (13 genera) Delostoma D. Don (2 species), Oroxyleae (1 genus), Tourrettia Foug. (2 species), Argylia D. Don (2 species), and Jacaranda Juss. (4 species) (Table S1). To determine the position of the genus Incarvillea within the family Bignoniaceae, we also used Pedalium murex L. and Sesamum indicum L. (only for genus-level phylogeny reconstruction) from the family Pedaliaceae (Gormley et al., 2015).
Total genomic DNA was extracted from dried leaves of four Incarvillea arguta specimens following the manufacturer's protocol for the DNAsecure Plant Kit (Tiangen Biotechnology Co. Ltd., Beijing, China). Plastid trnL-trnF sequences were obtained from Rana et al. (2019), whereas nrITS sequences were amplified using primers ITS1 and ITS4 (White et al., 1990). PCR reactions were performed in 30 μL volume containing 20–40 ng of sample DNA, 15 μL 2x Taq Plus Master Mix with dye (Tiangen Biotech), 12 μL of double distilled H2O and 1 μL of 10 μM stock of each primer. The reaction was performed under the following thermal conditions: 94 ℃ for 3 min, followed by 35 cycles at 94 ℃ for 30 s, 53 ℃ for 30 s and 72 ℃ for 7 min, with reactions held at 10 ℃ until further processing. PCR products were purified and sequenced by Tsingke Biological Technology Co. (Beijing, China).
2.2. Molecular phylogeny of the montane genus IncarvilleaPhylogenetic relationships of the genus Incarvillea within the family (family-level) and subgenera within the genus (genus-level) were inferred by using plastid trnL-trnF sequences. Phylogenetic estimates were based on Bayesian inference (BI), maximum likelihood (ML) and maximum parsimony (MP) approaches using MRBAYES v.3.2.6 (Ronquist et al., 2012), RAxML (Stamatakis, 2014) and PAUP∗ v.4.0a (Swofford, 2003) respectively. Furthermore, all three approaches were applied to reconstruct the subgeneric phylogenetic position within the genus Incarvillea using nrITS and combined (trnL-trnF and nrITS) sequences. Indels were treated as missing data, by default. We used GTR + G as the best fitting substitution model based on the Akaike information criterion (AIC) using JMODELTEST 2.1.6 (Posada and Crandall, 1998). Bayesian analysis was carried out in online CIPRES Science Gateway v.3.3 (Miller et al., 2010; https://www.phylo.org) using 5 × 107 generations, four chains with sampling every 1000th generations and first 20% was discarded as burn-in. ML analyses were performed using the graphical front-end raxmlGUIv1.5b2 (Silvestro and Michalak, 2012) for RAxML v.8.2.x with 1000 rapid bootstraps (Stamatakis, 2014). MP analyses were performed using heuristic searches with 1000 replicates of random addition, 10 trees held in each step during stepwise addition, tree bisection reconnection (TBR) swapping and 1000 bootstrapping replications with 10, 000 maximum number of trees. The bootstrap results are summarized in a 50% majority-rule consensus cladogram. The clade support on the tree shows the Bayesian posterior probabilities (PP), maximum likelihood bootstrap values and maximum parsimony bootstrap values estimated by BI, ML and MP approaches respectively. The degree of phylogenetic incongruence between trnL-trnF and ITS partitions as a combined sequence was assessed using the incongruence-length difference (ILD) test (Farris et al., 1994, 1995) in PAUP∗ v.4.0a, with 1000 replications using a heuristic tree search and 1000 additional sequence replicates. Furthermore, only parsimony-informative sites were used to ensure the accuracy of the P-value in the ILD test.
2.3. Estimation of divergence timeWe calculated the divergence time based on the fossil and secondary calibration points using Bayesian statistics in BEAST v.1.8.4 (Drummond and Rambaut, 2007; Drummond et al., 2012). The Bayesian relationship of plastid trnL-trnF datasets were conducted using a TVM + G substitution model selected by the AIC in JMODELTEST 2.1.6 (Posada and Crandall, 1998) and uncorrelated lognormal relaxed clock (P < 0.05, a likelihood-ratio test in PAUP∗ v.4.0a; Swofford, 2003). A yule process was specified as tree prior. Three calibration points were used to construct nodes (nodes 1, 2, and 3). First, based on meta-calibration work on flowering plants (Magallon et al., 2015), the divergence time between Bignoniaceae and Pedaliaceae (Pedalium murex) was set to 47.03 Ma as our prior for the root node 1. We assumed a normal distribution with a mean of 47.03 Ma, and a standard deviation of 5.8, giving a 95% HPD of 37.49–56.57 Ma. Several other fossil records reveal that the age of the family is ca. 50 million years old (e.g. Wehr and Hopkins, 1994; Wehr, 1995; Wehr and Manchester, 1996; Wilf, 1997; Pigg and Wehr, 2002). Lohmann et al. (2013) applied these fossil records using a normal prior with a mean of 49 Ma and a standard deviation of 3.0 to calibrate Bignoniaceae. Secondly, fossil records for the Catalpa–Campsis radicans, which are estimated as 38.8 Ma, sets as the minimum age for the clade (Meyer and Manchester, 1997; Manchester, 2000) to calibrate the crown node 2 of the Catalpa–Campsis clade. We used a lognormal distribution with the minimum age constraints 38.8 Ma taken as zero offset, and both the lognormal mean and the standard deviation set to 1 (95% HPD: 38.94–52.5 Ma). The fossil from the seed of Catalpa Scop. from the John Day Formation, Wheeler County, Oregon, USA (Meyer and Manchester, 1997), is estimated to be from the Late Eocene. The fossil-bearing sediments are estimated to be ca. 38.8 Ma, based on the dating of the overlying tuff (38.4 ± 0.7 Ma), and the underlying ignimbrite (39.17 ± 0.15 Ma; Manchester, 2000). Finally, the secondary calibration point estimated as 19.906 Ma was set as the divergence of the genus Incarvillea lineages (Rana et al., 2019). We assumed a normal distribution with a mean of 19.906 Ma, and a standard deviation of 3.89, giving a 95% HPD of 13.51–26.3 Ma as our prior for the crown node.
BEAST used a MCMC chain length of 5 × 107 generations, sampling every 1000th generations, following a burn-in of the initial 20% cycles. MCMC samples were checked in TRACER to confirm sampling adequacy and convergence of the chains to a stationary distribution. We started the analysis independently to detect the effective sample size (ESS > 200) for all parameters of each run (Rambaut and Drummond, 2007). TREEANNOTATOR v.1.8.4 (Drummond et al., 2012) and FIGTREE v.1.4.0 (Rambaut, 2009) were used to summarize and display the sampled trees, respectively.
2.4. Ancestral area reconstructionTo limit methodological biases from influencing subsequent inferences, we used multiple approaches to reconstruct the ancestral area of the genus Incarvillea. A dispersal-extinction-cladogenesis model (DEC-Lagrange) and Bayesian Binary Method (BBM) were implemented in LAGRANGE v.20130526 (Ree and Smith, 2008) and RASP v.4.0 (Reconstruct Ancestral State in Phylogenies; Yu et al., 2015), respectively, using all post-burn-in trees from the BEAST analysis. We excluded all the outgroups of BEAST MCMC trees from chronograms to represent only the biogeographic range of the genus Incarvillea. Biogeographic data for species within the genus Incarvillea were compiled from the distribution information described in the literature, online flora of China and herbarium specimens. We divided the entire geographic range of the genus Incarvillea (Fig. 1 a) into six areas corresponding to the floristic divisions of East Asia (Wu and Wu, 1996; Sun, 2013): (A) the Sino-Japanese region and Mongolian steppe region (SJM), (B) the Sino-Himalaya region (SH), (C) the QTP region, (D) the Western Himalaya (WH), (E) the Far-West Himalaya (FWH), and (F) Central Asia (CA). The number of maximum areas occupied at each node was set to four. For these analyses, range constraints were restricted to the region 'E and F' from 'A, B' and vice-versa. The dispersal rate between areas was set to 1 (without barrier; from A to B, C; B to C, D; C to D, E; E to F and vice versa); 0.5 (intermittent barriers; from A to D, E; B to E and vice versa) and 0.01 (with barriers; from A–B to F and vice versa) (Zhou et al., 2019). The BBM was run with the fixed state frequencies model (Jukes-Cantor) with equal among-site rate variation for 10 independent runs of 50, 000, 000 generations of the MCMC chains, with sampling every 1000th generations; the first 20% was discarded as burn-in in BBM analyses.
2.5. Stochastic character mappingWe created a matrix of 10 morphological characters that included habit, leaf arrangement, calyx teeth base, inflorescence, anther texture, capsule shape, capsule texture, capsule wing, capsule dehiscence, and seed wing (Table S2). The characters were selected based on Chen et al.(2005, 2006), the published literature, flora of China and examination of herbarium specimens, photographs (Table 1). We scored these characters for each species included in the plastid trnL-trnF data set, except for the outgroup taxa. Maximum likelihood reconstruction of ancestral states as a function of stochastic character mapping (SCM) (Huelsenbeck et al., 2003) was implemented to reconstruct the evolution of morphological characters of the genus Incarvillea. The package phytools v.0.6–99 (Revell, 2019) in R (R Core Team, 2019) was used to project the habitats onto the maximum clade credibility tree from a BEAST analysis. The best-fitting model for each character was selected by likelihood ratio tests, and the set of adequately fitting models was found by comparing corrected Akaike information criterion (AICc) scores. Out of the three transition models (ER, equal rates model; SYM, symmetrical model; ARD, all-rates-different model), we used ER following model selection via the corrected AICc (Table 3). The AICc is the Akaike information criterion for a finite sample size (Burnham and Anderson (2002) recommend the use of AICc rather than AIC when n/K < 40; n = sample size, K = number of degrees of freedom).
3. Results 3.1. Sequence data and phylogenetic relationshipsThe family-level phylogeny was represented by 75 trnL-trnF accessions (18 accessions of 13 species as ingroup and 57 accessions as outgroups). The aligned matrix was 1083 characters long with 208 phylogenetically parsimony-informative sites and 144 parsimony-uninformative sites. For both family and genus-level phylogenetic relationships, we compared the most likely tree obtained from the BI, ML and MP analyses, along with their support values. We considered weak support (0.5 < PP < 0.7), moderate support (0.7 < PP < 0.95) and strong support (PP > 0.95) for Bayesian posterior probabilities, whereas good support (70 < values < 95) and strong support (values > 95) for likelihood and parsimony analyses.
The phylogenetic trees generated by BI, ML and MP analyses were somewhat congruent, differing only in the support values of the nodes (Fig. 2). BI, ML and MP analyses strongly support the genus Incarvillea being a monophyletic clade within Tecomeae (PP = 1, values = 100) (Fig. 2, Fig. 3, Fig. 4 and S1–6). The phylogenetic framework of the genus Incarvillea based on plastid trnL-trnF in this study is consistent with a previous molecular study by Olmstead et al. (2009) (Fig. 2). The five clades within the genus Incarvillea are equivalent to the five subgenera (Amphicome, Olgaea, Niedzwedzkia, Incarvillea and Pteroscleris) as proposed by Grierson (1961) and Chen et al.(2005, 2006). Of the five major clades, three clades (subgenera Niedzwedzkia, Incarvillea, Pteroscleris) are rooted as one with strong support (Fig. 2, Fig. 3). It makes sense that the subgenus Niedwedzkia (the Central Asian species) followed by subgenus Incarvillea is sister to subgenus Pteroscleris (East Asian species) clade (Fig. 2, Fig. 3 a and S1), but with weak to moderate support. This topology was used to estimate divergence time, reconstruct the ancestral area and analyse character evolution.

|
| Fig. 2 Bayesian majority-rule consensus tree of the genus Incarvillea within Bignoniaceae (except Bignonieae) inferred from plastid trnL-trnF sequence. Bayesian posterior probabilities (PP > 0.50) are represented above the branches. Maximum likelihood bootstrap values/maximum parsimony bootstrap values are indicated below branches, with "–ˮ indicating support values of less than 50% or collapse in analysis. Colour legend for the genus Incarvillea clade: purple, subgenus Pteroscleris; blue, subgenus Incarvillea; cyan, subgenus Niedzwedzkia; green, subgenus Amphicome; red, subgenus Olgaea. |

|
| Fig. 3 Bayesian majority-rule consensus tree of the genus Incarvillea inferred from (a) plastid trnL-trnF, (b) nrITS, and (c) combined (trnL-trnF and nrITS) sequences. Bayesian posterior probabilities (PP > 0.50) are represented below the branches. Maximum likelihood bootstrap values/maximum parsimony bootstrap values are indicated above branches, with "–ˮ indicating support values of less than 50% or collapse in analysis. Colour legend for the genus Incarvillea clade: purple, subgenus Pteroscleris; blue, subgenus Incarvillea; cyan, subgenus Niedzwedzkia; green, subgenus Amphicome; red, subgenus Olgaea. |
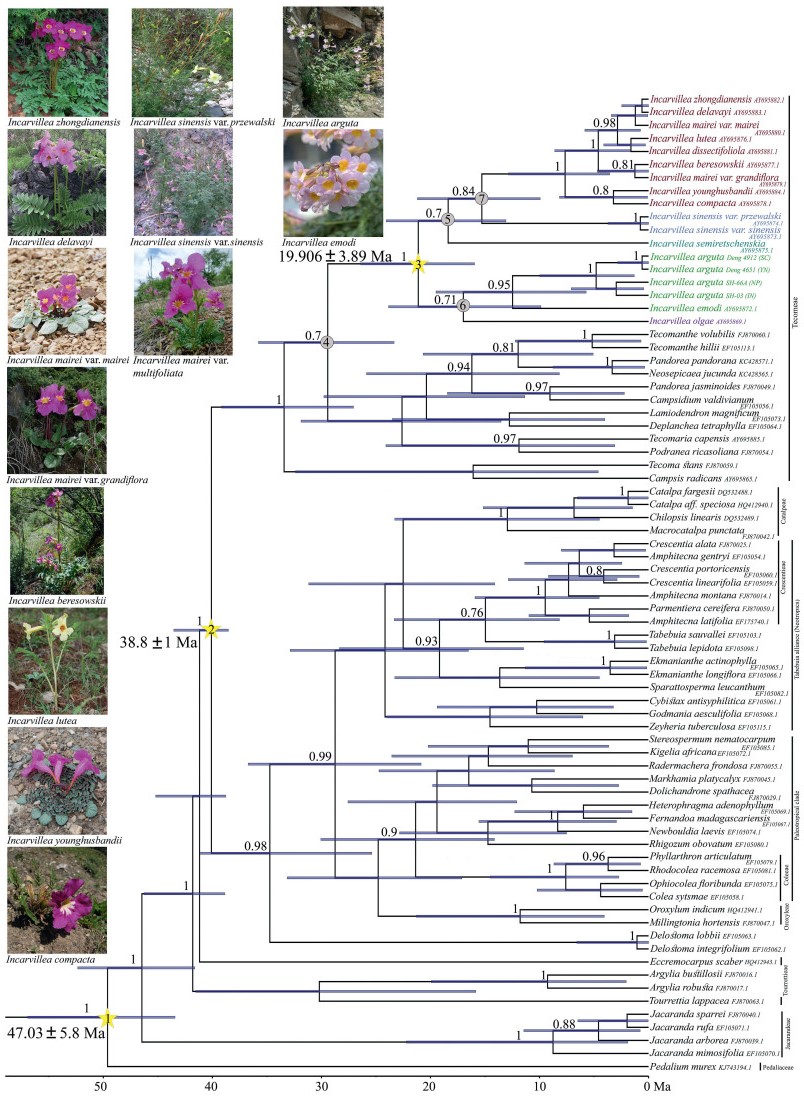
|
| Fig. 4 BEAST-derived chronogram of Bignoniaceae (except Bignonieae) to estimate the divergence time of the genus Incarvillea based on trnL-trnF sequences with calibration points denoted by nodes 1, 2, and 3 in star. Blue bars indicate the 95% highest posterior density (HPD) credibility intervals for node ages (in Ma). Bayesian posterior probabilities (> 50%) are sequentially labelled above the branch. The numbered nodes (4–7) are stem and crown nodes (see Table 2 for estimated age). (Photographs: Hang Sun: I. delavayi, I. compacta; Niu Yang: I. mairei; Shaotian Chen: I. beresowskii, I. lutea, I. sinensis, I. younghusbandii, I. zhongdianensis; Santosh Kumar Rana: I. arguta, I. emodi). |
Genus-level phylogenetic position was inferred separately using 22 accessions (18 accessions of 13 Incarvillea species and four outgroups) of each plastid trnL-trnF, nrITS and combined sequences. In the homogeneity test, the combined data sets provided a P value of 0.79: the acceptance of the null hypothesis. Therefore, the plastid trnL-trnF and nrITS datasets were combined into a single matrix for phylogenetic analyses. The aligned matrices for trnL-trnF, nrITS and combined sequences were 979, 761 and 1740 characters long with 86, 144 and 230 phylogenetically parsimony-informative sites, and 48, 78 and 126 parsimony-uninformative sites respectively. The two data sets used in the genus-level phylogeny also indicate that the genus Incarvillea is a monophyletic group and is sister to Tecomaria capensis (Thunb.) Spach (Fig. 3). These relationships were supported by 100% posterior probabilities and bootstrap values (Figs. 3a and S1), except when ML and MP analyses were used with the nrITS data set (PP = 0.70, bootstrap value = 84% by trnL-trnF) (Figs. 3b; S2 and S5). The five major clades equivalent to five subgenera as proposed by Chen et al. (2005) are resolved with moderate to high support values by trnL-trnF (Fig. 3). Of the five subgenera, subgenus Niedzwedzkia (Central Asian I. semiretschenskia Grierson) is rooted with East Asian subgenera Incarvillea and Pteroscleris with 80% support (Fig. 3 a). The interrelationships of the five major sub-generic clades were only moderately resolved; for instance, the sister–group relationship between I. sinensis (subgenus Incarvillea) and members of the subgenus Pteroscleris was ambiguously supported by ML/MP analyses (bootstrap values = 50/71.5) (Figs. 3a; S1 and S4). In contrast, nrITS and combined data sets revealed a concordance topology of the subgenera, differing in the support values and placement of I. semiretschenskia (subgenus Niedzwedzkia) (Figs. 3b and c; S2; S3; S5 and S6). The nrITS data sets fully resolved five sub-generic classifications, but lacked strong support for subgenera Olgaea and Amphicome. However, when combined data sets were used, subgenus Amphicome was robustly supported by BI (Fig. 3b) and moderately supported by ML/MP (Figs. S2 and S5). The combined data set analysis also had higher support values on the ingroup nodes than separate analyses, and generated moderately resolved support values for subgenus Olgaea (Figs. 3; S3 and S6).
3.2. Divergence time estimationThe MCC tree based on trnL-trnF coincides with the phylogenetic framework of BI, ML and MP analyses for the plastid DNA datasets at the family or genus-level (Fig. 2, Fig. 3, Fig. 4). Our molecular dating analyses (Fig. 4; Table 2) suggest that the genus Incarvillea might have evolved before the Miocene, during the mid-Oligocene ca. 29.42 Ma (95% HPD: 23.34–35.78 Ma) and the earliest diverging lineages in the genus Incarvillea arose during the early Miocene ca. 21.12 Ma (95% HPD: 15.96–26.4 Ma). Most of the major clades of subgenera in the genus Incarvillea were established before the Quaternary period. The recent lineage diversification of subgenus Pteroscleris occurred during the late Miocene ca. 7.65 Ma (95% HPD: 3.56–12.85 Ma). The tribe Tecomeae clade diverged during the early Oligocene ca. 33.43 Ma (95% HPD: 27.06–39.17 Ma) (Fig. 4; Table 2).
| SN | Major clades and numbered nodes | Age estimates (Ma) | Ancestral area reconstructions | |||
| Mean | 95% HPD | BBM (Prob.) | DEC-Lagrange (Prob.) | |||
| 1 | Node 4, Stem node of the genus Incarvillea | 29.42 | 23.34-35.78 | NA | NA | |
| 2 | Node 5, subgenera Niedzwedzkia–Incarvillea–Pteroscleris clade | 18.38 | 13.08-24.06 | F (76.6%) B (13.3%) |
B|F (25%) | |
| 3 | Node 6, subgenera Olgaea–Amphicome clade | 16.99 | 9.87-23.85 | F (83%) D (7.28%) |
D|F (28.5%) | |
| 4 | Node 7, subgenera Incarvillea–Pteroscleris clade | 15.29 | 9.94-21.2 | B (83.5%) AB (4.6%) |
B|B (54%) | |
| 5 | Node 8, subgen. Pteroscleris clade | 7.65 | 3.56-12.85 | BC (53%) B (43%) |
B|B (37.1%) | |
| 6 | Tribe Tecomeae clade | 33.43 | 27.06-39.17 | NA | NA | |
| BMM, Bayesian binary Markov chain Monte Carlo; HPD, highest posterior density; DEC, Dispersale-Extinctione-Cladogenesis; Prob., relative probability. | ||||||
The result of DEC-Lagrange and BBM analyses is shown in Fig. 5 and interpreted in Table 2. Both analyses were congruent with most clades and indicated that the genus Incarvillea originated in Central Asia (area F; Fig. 5, node 3; DEC-Lagrange 30.7% and BBM 89.5%). Furthermore, the analyses indicated that species of the Central Asian genus Incarvillea may have dispersed to the Himalaya and the Mountains of SW China. For example, our analyses indicate that plants in the subgenus Pteroscleris rapidly diversified in the Sino-Himalaya after colonizing the Sino-Japanese and Mongolian steppe regions, the Western Himalaya, and the Qinghai-Tibet Plateau. The split between the subgenera Pteroscleris and Incarvillea was accompanied by the colonization of the Sino-Himalaya region, and Sino-Japanese and Mongolian steppe regions, along with the early divergence of subgenus Niedzwedzkia (see clade 5 of subgenera Niedzwedzkia–Incarvillea–Pteroscleris). Likewise, the colonization of the Sino-Himalaya and Far/Western Himalaya gave rise to the subgenus Amphicome, whose center of origin is the Western Himalaya (Rana et al., 2019) and is rooted with subgenus Olgaea from Central Asia. In general, results from DEC-Lagrange and BBM are similar to what is known about the biogeography of the genus Incarvillea. One notable difference, however, is that DEC-Lagrange suggested that lineages from the subgenus Pteroscleris colonized from the Sino-Himalaya only, whereas BBM indicated that colonization occurred from both the QTP and Sino-Himalaya.
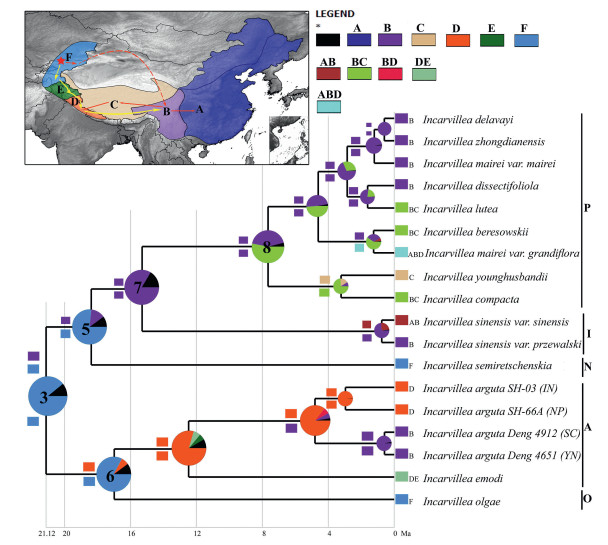
|
| Fig. 5 Ancestral area reconstruction of the genus Incarvillea based on the Bayesian Binary Markov Chain Monte Carlo (BBM), and dispersal–extinction–cladogenesis model (DEC-Lagrange) method implemented in RASP and Lagrange, respectively, using the BEAST-derived cpDNA chronogram. Ancestral areas shown by coloured boxes at nodes, where pie charts for each node inferred by BBM (at the node), and squared coloured box above and below the branch inferred by DEC-Lagrange illustrate the likelihood proportions and probabilities of each ancestral range. The colour key at outgroups corresponds to possible ancestral ranges. Putative biogeographic events are shown in upper left corner map. The different biogeographic regions represented by uppercase letters are shown on the map. A, Sino-Japanese and Mongolian steppe region (SJM); B, Sino-Himalaya region (SH, or Mountains of SW China); C, the QTP region; D, Western Himalaya (WH); E, Far-West Himalaya (FWH); and F, Central Asia (CA). Five subgenera are indicated by their initial letters. |
The likelihood ratio tests and the set of adequately fitting models obtained by comparing AICc scores indicated that ER and SYM were equally predictive for habit, calyx teeth base, inflorescence, anther texture, capsule shape, capsule wing, capsule dehiscence, whereas the remaining characters were favoured by ER model alone (Table 3). Therefore, to infer consistency of character mapping, we implemented the ER model as the best fitting model. The posterior probabilities inferred as the state frequencies calculated across 1000 stochastically mapped character histories for every 17 nodes (Table S2) were good enough to map characters using the ER model. Stochastic character mapping indicated that the ancestral characters of the genus Incarvillea include a suffruticose habit, alternate leaf arrangement, calyx teeth base not swollen, racemose inflorescence type, glabrous anther texture, cylindrical capsule shape, subligneous capsule texture, absence of capsule wing, loculicidal capsule dehiscence, and opaque seed wing (Fig. 1, Fig. 6). These characters are retained at the earliest diverging ancestral node across the genus. These character states persist through the backbone nodes with branches leading to several clades containing species of different subgenera. Most of the subgenus nodes share the ancestral characters except for anther texture, which is most commonly pilose (Fig. 1, Fig. 6e). In addition, the subgenus Amphicome has the highest likelihood of having fibrous capsules (Fig. 6g) and seed wing comas at the ends (Fig. 6j), but lack these ancestral characters.
| Character | Model | lnL | AICc | dAICc | wAICc |
| Habit | ARD | −3.5849 | 11.169853 | 1.61901 | 0.18203 |
| SYM | −3.7754 | 9.550848 | 0 | 0.40898 | |
| ER | −3.7754 | 9.550848 | 0 | 0.40898 | |
| Leaf arrangement | ARD | −9.1369 | 30.27383 | 6.54656 | 0.02992 |
| SYM | −10.341 | 26.6812 | 2.95393 | 0.18033 | |
| ER | −10.864 | 23.72727 | 0 | 0.78976 | |
| Calyx teeth base | ARD | −2.7632 | 9.526412 | 1.13493 | 0.22087 |
| SYM | −3.1957 | 8.391486 | 0 | 0.38957 | |
| ER | −3.1957 | 8.391486 | 0 | 0.38957 | |
| Inflorescence | ARD | −2.585 | 9.16992 | 1.08775 | 0.22495 |
| SYM | −3.0411 | 8.082168 | 0 | 0.38752 | |
| ER | −3.0411 | 8.082168 | 0 | 0.38752 | |
| Anther texture | ARD | −3.9023 | 11.804559 | 1.8105 | 0.16821 |
| SYM | −3.997 | 9.994056 | 0 | 0.4159 | |
| ER | −3.997 | 9.994056 | 0 | 0.4159 | |
| Capsule shape | ARD | −2.5665 | 9.133063 | 1.19576 | 0.21568 |
| SYM | −2.9687 | 7.937306 | 0 | 0.39216 | |
| ER | −2.9687 | 7.937306 | 0 | 0.39216 | |
| Capsule texture | ARD | −6.9027 | 25.80548 | 8.84777 | 0.01005 |
| SYM | −7.1922 | 20.38433 | 3.42662 | 0.1512 | |
| ER | −7.4789 | 16.95771 | 0 | 0.83875 | |
| Capsule wing | ARD | −2.5665 | 9.133063 | 1.19576 | 0.21568 |
| SYM | −2.9687 | 7.937306 | 0 | 0.39216 | |
| ER | −2.9687 | 7.937306 | 0 | 0.39216 | |
| Capsule dehiscence | ARD | −2.5665 | 9.133063 | 1.19576 | 0.21568 |
| SYM | −2.9687 | 7.937306 | 0 | 0.39216 | |
| ER | −2.9687 | 7.937306 | 0 | 0.39216 | |
| Seed wing | ARD | −8.0014 | 28.00270 | 8.84776 | 0.01005 |
| SYM | −8.2908 | 22.58155 | 3.42661 | 0.1512 | |
| ER | −8.5775 | 19.15494 | 0 | 0.83875 | |
| lnL, log-likelihood of the model; AICc, corrected Akaike information criterion; dAICc, the difference between the AICc of the model and the best model; wAICc, AICc weight of the model. The dAICci, AICci – min AICc follows the following criteria: dAICci ≤ 1, substantial support for a model with AICci; 4 ≤ dAICci ≤ 9, low support for a model with AICci; dAICci > 10, no support for a model with AICci. The AICc weights indicate the probability that the model is the best among the whole set of candidate models (Burnham and Anderson, 2002; Sullivan and Joyce, 2005). ER, equal rates model; SYM, symmetrical model; and ARD, all-rates-different model. Bold font in the Model columns indicates the selected model by the likelihood ratio test. | |||||
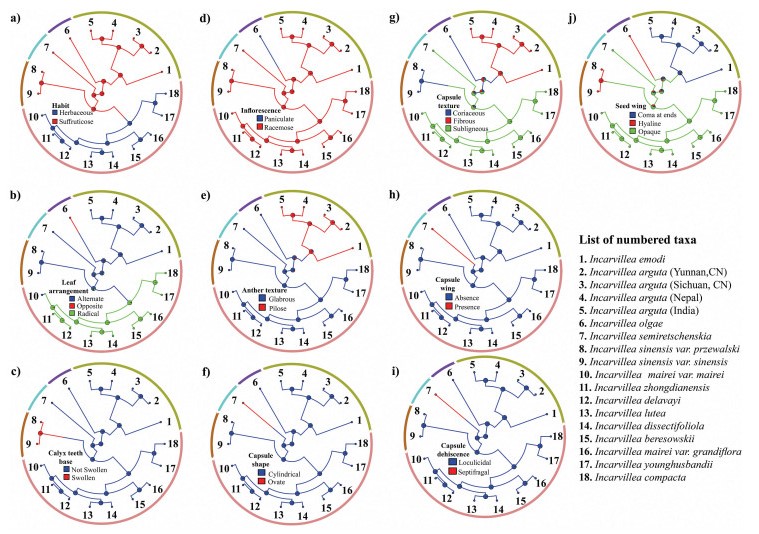
|
| Fig. 6 Maximum likelihood estimates on ancestral states of stochastic characters for the genus Incarvillea grouped into five subgenera based on best-fitting evolutionary transition models 'equal rate' (ER) depicted on the expanded chronogram in Fig. 5. Five subgenera are labelled and indicated in colour: green, subgenus Amphicome; blue, subgenus Olgaea; cyan, subgenus Niedzwedzkia; magenta, subgenus Incarvillea; red, subgenus Pteroscleris. The 13 taxa are numbered. Each stochastic character is quoted within the phylogram. Probabilities of states at ancestral nodes indicated by pie charts. |
The relationships between the five subgenera (Amphicome, Incarvillea, Pteroscleris, Olgaea and Niedawedzkia) of the genus Incarvillea have long remained unresolved. Previous studies that examined these relationships using either molecular data or morphological characters (habit, calyx, capsule, and seed) have generated inconsistent phylogenies (Grierson, 1961; Chen et al., 2005, 2006). In this study, phylogenetic analyses using trnL-trnF data sets indicate that the genus Incarvillea is monophyletic. Our phylogenies based on nrITS and combined data sets (Fig. 3b and c) are generally consistent with the generic classification of previous molecular studies by Chen et al. (2005). Our findings support the placement of subgenus Niedzwedzkia rooted within the subgenera Incarvillea and Pteroscleris, which contrasts with previous placements based on trnL-trnF sequences (Chen et al., 2005). Moreover, our molecular phylogeny based on trnL-trnF sequences revealed two major basal clades (i.e., Olgaea–Amphicome and Niedzwedzkia–Incarvillea–Pteroscleris), which also differs from Chen et al.'s phylogeny (2005). Within the second basal clade, the phylogenetic arrangement of the basal subgenus Niedzwedzkia (Grierson, 1961; Chen et al., 2005) resulted in more resemblances with subgenus Incarvillea. For example, the Central Asia endemic species I. semiretschenskia has the East Asian habitat type (Illarionova, 2006) and stalked glands identical to members of subgenus Incarvillea. The placement of taxa in subgenus Pteroscleris, as revealed by ML and BI, might differ due to their respective support values. These differences should be considered carefully within a phylogenetic framework as they indicate the evolutionary significance of these taxonomic relationships. Importantly, the phylogenetic positions of subgenera should be resolved using multilocus data. Here, we provide the separate subgeneric rank of the genus Incarvillea from two major basal clades revealed by trnL-trnF based Bayesian and Likelihood chronograms as: (((Olgaea (Amphicome)), (Niedzwedzkia, ((Incarvillea), (Pteroscleris)))). Although our findings differ to some extent from Grierson (1961), Flora Reipublicae Sinicae (Wang et al., 1990), and Chen et al. (2005), we believe that the subgeneric positions are reasonable based on morphology and biogeography. Therefore, correct placement of taxa requires that researchers consider morphological characters along with molecular data.
4.2. Geographic origin, dispersal and colonization of the montane genus Incarvillea within the three major biodiversity hotspotsOur biogeographic reconstruction proposes that the genus Incarvillea originated in Central Asia before the Miocene (i.e., during mid-Oligocene ca. 29.42 Ma) and subsequently dispersed to the Western Himalaya and Sino-Himalaya during the early Miocene (21.12 Ma) (95% HPD: 15.96–26.4 Ma) (Fig. 4, Fig. 5; Table 2). Although the genus may have originated in Central Asia, vicariance events in the Himalaya-Hengduan Mountains (HHM) may have led to allopatric speciation, creating differentiation centres or centres of endemism (Chen et al., 2005; Sun et al., 2017) (Fig. 5). The dispersal of biota between the Western Himalaya and the Sino-Himalaya is one of the most noteworthy events related to uplift of the Himalaya-Hengduan Mountains. Thus, successive dispersal and vicariance events within the Himalaya may have driven additional species divergence and lineage diversification. The lack of differentiation centres in the Western Himalaya may be related to the influence of 'Westerlies', prevailing winds from west to east, which enhance dispersal ability despite strong mountain barriers (Rana et al., 2019). This suggests that despite barriers to migration, weak dispersal of Incarvillea species might have been expedited by the Westerlies, aiding migration from their region of origin over long distances in the early Pliocene (Chen et al., 2005; Qiong et al., 2017).
Despite gaps in taxon sampling and uneven geographical sampling, previous studies suggest that the southern part of the QTP may provide suitable landscape connectivity for species to migrate (Rana et al., 2019). Mountains do not form uniform barriers; river valleys cut deep into the Himalayas, crossing drainage divides, and forming dispersal corridors. For members of Incarvillea, geoclimatic factors are also likely to influence species that show weak dispersal abilities (Rana et al., 2019) as revealed by Rana et al. (2020a) in Mirabilis himalaica (Edgew.) Heimerl. Pollen dispersal in Incarvillea species is mediated by specialized floral structures with obvious herkogamy, often including a bilobed sensitive stigma and anther appendages (Cutting, 1921; Han et al., 2008; Ai et al., 2013). However, the reproductive strategies of alpine species of the genus Incarvillea endemic to the HHM region have not yet been fully characterized (Ai et al., 2013). Moreover, temperature shifts in mountain environments possess strong effects on the elevational distributions of species (Pearson and Dawson, 2003), limiting further suitable expansion. Therefore, plants must either adapt to existing environments and colonize longitudinally within the environment or suffer extinction (Rana et al., 2018, 2019, 2020a, b).
A floristic relationship between Central Asian and East Asian Incarvillea is likely to exist. The connection is supported by the distribution of the Central Asian endemic I. semiretschenskia, which is found outside of the East Asia type habitat in the eastern part of the ancient Chu-Ili mountains (Illarionova, 2006). Such a connection can also be seen when the distribution range of the genus Incarvillea is projected under different climatic scenarios (paleo-climate, current, future), forming a ring of suitability around the QTP (Rana et al. unpublished manuscript). Our study may have found support for the biogeographic connection of the Central Asian species Incarvillea semeretscheskia within the subgenera Incarvillea and Pteroscleris if we had sampled I. potaninii from Mongolia. Nevertheless, the species I. potaninii of subgenus Incarvillea is still distributed in Mongolia (north of the QTP), which suggests that a floristic relationship between Central Asia and East Asia exists.
Alternatively, biogeographic analyses using Incarvillea potaninii (from Mongolia) may indicate that the dispersal route between Central and East Asia was direct rather than through the Mountains of SW China, which strongly supports the sister–group relationship of subgenus Niedzwedzkia with subgenera Incarvillea and Pteroscleris. The continuous distribution of the genus Incarvillea from the Mountains of Central Asia, the Himalaya-Hengduan Mountains to NE China and Mongolia, may have partly resulted from the Quaternary expansion as part of its range was glaciated (Chen et al., 2005).
Regardless, the intense dispersal of Incarvillea occurred from the Miocene onwards, while the colonization of the biodiversity hotspots and adjoining regions by representatives of Incarvillea subgenera started during the Quaternary period. The accelerated floristic exchange between the three major biodiversity hotspots (along with adjoining regions) might have been encouraged by Miocene cooling (Flower and Kennett, 1994; Zachos et al., 2001), which increased the occurrence of montane habitats in these regions.
4.3. Morphological characters evolutionWe inferred ancestral states for several taxonomically important morphological stochastic characters to nodes deeply nested within the tree (Fig. 6). Our study strongly confirms that the most precise ancestral character that categorizes the genus Incarvillea is suffruticose habit. Morphological data has traditionally been used to differentiate between Incarvillea subgenera. Of the characters we tested, capsule texture and seed wing, which had the highest AICc weights of the model, showed the greatest ability to differentiate between the five Incarvillea subgenera (Chen et al., 2004).
Additional specific ancestral characters distinguish Incarvillea subgenera. The large subgenus Pteroscleris is characterized by herbaceous habit and radical leaves, which are derived from ancestral suffruticose habit and alternate leaves, respectively. Subgenus Pteroscleris also shares the following characters with subgenus Incarvillea: racemose inflorescence, glabrous anther, cylindrical capsule, absence of capsule wing, and loculicidal capsule dehiscence. Taken together, these findings indicate that subgenus Pteroscleris is advanced and is more closely related to subgenus Incarvillea (Grierson, 1961; Chen et al., 2005). The characters that typify the subgenus Amphicome are primitive (Grierson, 1961; Chen et al., 2005), except for the frequency with which they express the ancestral states of some characters (e.g., pilose anther texture, fibrous capsule texture, commas at the end of seed wings). These characteristics are consistent with Grierson's views of the evolution of these three subgenera (Chen et al., 2005), suggesting that the subgenus Amphicome is ancestral.
An alternative interpretation is that the subgenus Amphiocme is primitive, subgenus Pteroscleris is advanced, and subgenus Incarvillea is an intermediate between these two genera (Chen et al., 2004, 2005). However, the two other subgenera (i.e., Niedzwedzkia and Olgaea) that form the two major lineages diverged 21.12 Ma, which would place the subgenus Amphicome at the root with Olgaea, and the subgenera Pterocleris and Incarvillea at the root with subgenus Niedzwedzkia. The two subgenera (Olgaea and Niedzwedzkia) that diverged early share ancestral character states. Specifically, the subgenus Niedzwedzkia, which diverged first (18.38 Ma), possesses the most ancestral character states (e.g., suffruticosa habit, alternate leaves, a calyx teeth base that is not swollen, racemose inflorescence, glabrous anthers, subligneous capsule texture and opaque seed wings).
Incarvillea semiretschenskia (subgenus Niedzwedzkia) is the only species with a septifragal capsule (Chen et al., 2005) and possibly could have maintained the ancestral state in term of fruit dehiscence with a basal position in the genus. But, the formation of two main lineages in the genus could have resulted in the loculicidal capsule dehiscence as the ancestral state from subgenus Olgaea, from which the septifragal dehiscence might be derived. It seems plausible that not only the capsule dehiscence but also the cylindrical capsule shape and absence of capsule wing from the subgenus Olgaea are the ancestral state. The morphologically varied and distinct capsule characteristics (e.g., cylindric, winged and septifragal dehiscence) are peculiar evolutionarily derived character state. Hence, the subgenus Incarvillea does not appear to form an intermediate between subgenera Pteroscleris and Amphicome, but instead forms a transition between subgenera Pteroscleris and Niedzwedzkia. Additional evidence from the East Asian I. semiretschenskia, an endemic to Central Asia (Kazakhstan) with stalked glands, may support this hypothesis (Vassilczenko, 2000; Illarionova, 2006).
5. ConclusionsOur study re-examines the phylogenetic framework of the genus Incarvillea using trnL-trnF and nrITS (individually), and combined molecular data sets by BI, ML and MP. Furthermore, this study represents the first step towards an understanding of the biogeographical history of the genus Incarvillea. Our time-calibrated phylogenetic tree revealed that the genus Incarvillea is monophyletic and originated in Central Asia during mid-Oligocene ca. 29.42 Ma (95% HPD: 23.34–35.78 Ma) and the earliest diverging lineages arose during the early Miocene ca. 21.12 Ma (95% HPD: 15.96–26.4 Ma). Diversification of these lineages followed two simultaneous paths: dispersal to the Himalaya led to the Olgaea–Amphicome clade; dispersal to the Hengduan Mountains led to the Niedzwedzkia–Incarvillea–Pteroscleris clade. The subgenus Pteroscleris may have undergone a recent adaptive radiation, as it is a relatively new colonizer. Although our ability to make biogeographical inferences is limited due to low bootstrap support for nodes and limited taxon sampling, these analyses provide general insights into the complex history of the Incarvillea genus, which is characterized by repeated colonization of biogeographically defined areas. Stochastic character mapping provides strong evidence that there are two major lineages with ancestral character states; however, the phylogenetic placement of subgenus Niedzwedzkia must await further clarification. In the meantime, we expect that work on character evolution and the phylogeny of the genus Incarvillea will be well served by placing subgenus Niedzwedzkia close to subgenus Incarvillea. Finally, although base-line taxonomic work and additional sample collection are required, this study demonstrates the usefulness of using large secondary molecular data sets and biogeography to determine the evolutionary origins and diversification of plant taxa that occur in biodiversity hotspots.
Author contributionsSKR, SC and HS conceived the research design. SKR, HKR and DL performed the data analysis. SKR wrote the manuscript with the support of DL, HKR, and HS. All authors contributed to the final editing.
Declaration of competing interestTo the best of our knowledge, the named authors have no conflict of interest, financial or otherwise.
AcknowledgementsThe authors thank Dr Zhuo Zhou for his help with the molecular dating analysis, Raymond Porter and Alexander Robert O'Neill (USA) for valuable inputs and English editing, and Dr Niu Yang, Ya-Zhou Zhang, Qia Wang for providing the plant photographs. This study was supported by the Second Tibetan Plateau Scientific Expedition and Research (STEP) Program (2019QZKK0502), the Strategic Priority Research Program of Chinese Academy of Sciences (XDA20050203), NSFC-Yunnan joint fund to support key projects (U1802242), the Major Program of the National Natural Science Foundation of China (31590823) and the National Natural Science Foundation of China (31570203).
Appendix A. Supplementary dataSupplementary data to this article can be found online at http://www.R-project.org/.
Ai H.L., Zhou W., Xu K., et al, 2013. The reproductive strategy of a pollinatorlimited Himalayan plant, Incarvillea mairei (Bignoniaceae). BMC Plant Biol, 13: 195. DOI:10.1186/1471-2229-13-195 |
Burnham K.P., Anderson D.A., 2002. Model Selection and Multimodel Inference: a Practical Information-Theoretic Approach, second ed. New York: Springer-Verlag,.
|
Chen S.T., Gong J., Guan K.Y., et al, 2010. Biodiversity conservation of the genus Incarvillea Juss. (Bignoniaceae) based on molecular diversity and species richness assessment. J. Plant Biol, 53(6): 387-394. DOI:10.1007/s12374-010-9127-6 |
Chen S.T., Guan K.Y., Zhou Z.K., 2006. A new subgenus of Incarvillea (Bignoniaceae). Ann. Bot. Fenn, 43: 288-290. |
Chen S.T., Guan K.Y., Zhou Z.K., et al, 2005. Molecular phylogeny of Incarvillea(Bignoniaceae) based on ITS and trnL-F sequences. Am. J. Bot, 92(4): 625-633. DOI:10.3732/ajb.92.4.625 |
Chen S.T., Xing Y.W., Su T., et al, 2012. Phylogeographic analysis reveals significant spatial genetic structure of Incarvillea sinensis as a product of mountain building. BMC Plant Biol, 12: 58. DOI:10.1186/1471-2229-12-58 |
Chen S.T., Zhou Z.K., Guan K.Y., et al, 2004. Karyomorphological study of Incarvillea Juss. (Bignoniaceae) and its implications in distribution and taxonomy. Bot. J. Linn. Soc, 144(1): 113-121. DOI:10.1111/j.0024-4074.2004.00189.x |
Cutting E.M., 1921. On the pollination mechanism of Incarvillea delavayi Franch. Ann. Bot, 35(1): 63-71. DOI:10.1093/oxfordjournals.aob.a089748 |
Deng T., Chen Y.S., Wang H.C., et al, 2018. Molecular phylogeny and biogeography of Adenocaulon highlight the biogeographic links between new world and old world. Front. Ecol. Evol, 5: 162. DOI:10.3389/fevo.2017.00162 |
Drummond A.J., Rambaut A., 2007. BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evol. Biol, 7: 214. DOI:10.1186/1471-2148-7-214 |
Drummond A.J., Suchard M.A., Xie D., et al, 2012. Bayesian phylogenetics with BEAUti and the BEAST1. 7. Mol. Biol. Evol, 29(8): 1969-1973. DOI:10.1093/molbev/mss075 |
Farris J.D., Kallersjo M., Kluge A.G., et al, 1994. Testing significance of incongruence. Cladistics, 10: 315-319. DOI:10.1111/j.1096-0031.1994.tb00181.x |
Farris J.D., Kallersjo M., Kluge A.G., et al, 1995. Constructing a significance test for incongruence. Syst. Biol, 44: 570-572. DOI:10.2307/2413663 |
Filatov D.A., Osborne O.G., Papadopulos A.S.T., 2016. Demographic history of speciation in a Senecio altitudinal hybrid zone on Mt. Etna. Mol. Ecol, 25(11): 2467-2481. DOI:10.1111/mec.13618 |
Flower B.P., Kennett J.P., 1994. The Middle Miocene climatic transition: east Antarctic ice sheet development, deep ocean circulation and global carbon cycling. Palaeogeogr. Palaeoclimatol. Palaeoecol, 108(3-4): 537-555. DOI:10.1016/0031-0182(94)90251-8 |
Gaston K.J., 2000. Global patterns in biodiversity. Nature, 405: 220-227. DOI:10.1038/35012228 |
Gormley I.C., Bedigian D., Olmstead R.G., 2015. Phylogeny of Pedaliaceae and martyniaceae and the placement of trapella in Plantaginaceae s. l. Syst. Bot, 40(1): 259-268. DOI:10.1600/036364415X686558 |
Grierson A.J.C., 1961. A revision of the genus Incarvillea. Notes R. Bot. Gard. Edinb, 23: 303-354. |
Grose S.O., Olmstead R.G., 2007. Evolution of a charismatic neotropical clade: molecular phylogeny of Tabebuia s. l., Crescentieae, and allied genera (Bignoniaceae). Syst. Bot, 32(3): 650-659. DOI:10.1600/036364407782250553 |
Guo Z.T., Ruddiman W.F., Hao Q.Z., et al, 2002. Onset of Asian desertification by 22 Myr ago inferred from loess deposit in China. Nature, 416: 159-163. DOI:10.1038/416159a |
Hajra P.K., Rao R.R., 1990. Distribution of vegetation types in northwest Himalaya with brief remarks on phytogeography and floral resource conservation. Proc. Indian Acad. Sci, 100(4): 263-277. DOI:10.1007/BF03053480 |
Han Y., Dai C., Yang C.F., et al, 2008. Anther appendages of Incarvillea trigger a pollen-dispensing mechanism. Ann. Bot, 102(3): 473-479. DOI:10.1093/aob/mcn102 |
Harrison T.M., Copeland P., Kidd W.S.F., et al, 1992. Raising tibet. Science, 255(5052): 1663-1670. DOI:10.1126/science.255.5052.1663 |
Huelsenbeck J.P., Nielsen R., Bollback J.P., 2003. Stochastic mapping of morphological characters. Syst. Biol, 52(2): 131-158. DOI:10.1080/10635150390192780 |
Illarionova, I.D., 2006. Plants of Central Asia, vol. 15, pp. 6-11. Moscow.
|
Li G.D., Kim C., Zha H.G., et al, 2014. Molecular phylogeny and biogeography of the arctic-alpine genus Lagotis (Plantaginaceae). Taxon, 63(1): 103-115. DOI:10.12705/631.47 |
Li J.J., Shi Y.F., Li B.Y., 1995. Uplift of the Qinghai-Xizang (Tibet) Plateau and Global Change.. Lanzhou: Lanzhou University Press.
|
Liu J.Q., Gao T.G., Chen Z.D., et al, 2002. Molecular phylogeny and biogeography of the Qinghai-Tibet Plateau endemic Nannoglottis (Asteraceae). Mol.Phylogenet. Evol, 23(3): 307-325. DOI:10.1016/s1055-7903(02)00039-8 |
Lohmann L.G., Bell C.D., Calio M.F., et al, 2013. Pattern and timing of biogeographical history in the Neotropical tribe Bignonieae (Bignoniaceae). Bot. J. Linn. Soc, 171(1): 154-170. DOI:10.1111/j.1095-8339.2012.01311.x |
Lopez-Pujol J., Zhang F.M., Sun H.Q., et al, 2011. Mountains of southern China as "plant Museums" and "plant cradles": evolutionary and conservation insights. Mt. Res. Dev, 31(3): 261-269. DOI:10.1659/MRD-JOURNAL-D-11-00058.1 |
Magallon S., Gomez-Acevedo S., Sanchez-Reyes L.L., et al, 2015. A metacalibrated time-tree documents the early rise of flowering plant phylogenetic diversity. New Phytol, 207(2): 437-453. DOI:10.1111/nph.13264 |
Manchester S., 2000. Late eocene fossil plants of the John Day Formation, wheeler county, Oregon. Oregon Geol, 62(3): 51-63. |
Meyer H., Manchester S., 1997. The Oligocene Bridge Creek Flora of the John Day Formation, Oregon.. Los Angeles & London: University of California Pres.
|
Miller, M.A., Pfeiffer, W., Schwartz, T., 2010. Creating the CIPRES Science Gateway for inference of large phylogenetic trees. In: Proceedings of the Gateway Computing Environments Workshop (GCE). New Orleans, LA, pp. 1-8.
|
Milne R.J., Abbott R.J., 2002. The origin and evolution of Tertiary relict floras. Adv.Bot. Res, 38: 281-314. DOI:10.1016/S0065-2296(02)38033-9 |
Mittermeier, R.A., Turner, W.A., Larsen, F.W., et al., 2011. Global biodiversity conservation: the critical role of hotspots. In: Zachos, F., Habel, J. (Eds. ), Biodiversity Hotspots. Springer, Berlin, pp. 3-22.
|
Myers N., Mittermeier R.A., Mittermeier C.G., et al, 2000. Biodiversity hotspots for conservation priorities. Nature, 406: 853-858. DOI:10.1038/35002501 |
Nauheimer L., Boyce P.C., Renner S.S., 2012. Giant taro and ITS relatives: a phylogeny of the large genus Alocasia (Araceae) sheds light on Miocene floristic exchange in the Malesian region. Mol. Phylogenet. Evol, 63(1): 43-51. DOI:10.1016/j.ympev.2011.12.011 |
Nie Z.L., Sun H., Beardsley P.M., et al, 2006. Evolution of biogeographic disjunction between eastern Asia and eastern north America in Phryma (Phrymaceae). Am. J. Bot, 93(9): 1343-1356. DOI:10.3732/ajb.93.9.1343 |
Olmstead R.G., Zjhra M.L., Lohmann L.G., et al, 2009. A molecular phylogeny and classification of Bignoniaceae. Am. J. Bot, 96(9): 1731-1743. DOI:10.3732/ajb.0900004 |
Pearson R.G., Dawson T.P., 2003. Predicting the impacts of climate change on the distribution of species: are bioclimate envelope models useful? Global Ecol. Biogeogr, 12(5): 361-371. DOI:10.1046/j.1466-822X.2003.00042.x |
Pigg K.B., Wehr W.C., 2002. Tertiary flowers, fruits, and seeds of Washington State and adjacent areas-part Ⅲ. Wash. Geol, 30: 3-20. |
Posada D., Crandall K.A., 1998. MODELTEST: testing the model of DNA substitution. Bioinformatics, 14(9): 817-818. DOI:10.1093/bioinformatics/14.9.817 |
Qiong L., Zhang W.J., Wang H., et al, 2017. Testing the effect of the Himalayan mountains as a physical barrier to gene flow in Hippophae tibetana Schlect. (Elaeagnaceae). PloS One, 12(5): e0172948. DOI:10.1371/journal.pone.0172948 |
R Core Team, 2019. R: a Language and Environment for Statistical Computing. R Foundation for statistical computing, Austria. http://tree.bio.ed.ac.uk/software/FigTree/.
|
Rambaut, A., 2009. FigTree Version 1.3.1. Institute of Evolutionary Biology, University of Edinburgh. http://beast.bio.ed.ac.uk/Tracer. (Accessed 20 January 2019).
|
Rambaut, A., Drummond, A.J., 2007. Tracer Version 1.4. Molecular Evolution, Phylogenetics and Epidemiology. https://doi.org/10.3389/fpls.2019.01721. (Accessed 20 January 2019).
|
Rana S.K., Luo D., Rana H.K., et al, 2019. Geoclimatic factors influence the population genetic connectivity of Incarvillea arguta (Bignoniaceae) in the HimalayaeHengduan Mountains biodiversity hotspot. J. Syst. Evol, 1111. DOI:10.1111/jse.12521 |
Rana, H.K., Luo, D., Rana, S.K., et al., 2020a. Geological and climatic factors affect the population genetic connectivity in Mirabilis himalaica (Nyctaginaceae): insight from phylogeography and dispersal corridors in the Himalaya-Hengduan Biodiversity Hotspot. Front. Plant Sci. 10, 1721. https://doi.org/10.3389/fpls.2019.01721.
|
Rana S.K., Rana H.K., Ranjitkar S., et al, 2020b. Climate-change threats to distribution, habitats, sustainability and conservation of highly traded medicinal and aromatic plants in Nepal. Ecol. Indic, 115: 106435. DOI:10.1016/j.ecolind.2020.106435 |
Rana S.K., Rana H.K., Shrestha K.K., et al, 2018. Determining bioclimatic space of Himalayan alder for agroforestry systems in Nepal. Plant Divers, 40(1): 1-18. DOI:10.1016/j.pld.2017.11.002 |
Ree R.H., Smith S.A., 2008. Maximum likelihood inference of geographic range evolution by dispersal, local extinction, and cladogenesis. Syst. Biol, 57(1): 4-14. DOI:10.1080/10635150701883881 |
Refulio-Rodriguez N.F., Olmstead R.G., 2014. Phylogeny of lamiidae. Am. J. Bot, 101(2): 287-299. DOI:10.3732/ajb.1300394 |
Revell, L.J., 2019. Phytools: Phylogenetic Tool for Comparative Biology (And Other Things). https://cran.r-project.org/web/packages/phytools/phytools.pdf.
|
Ronquist F., Teslenko M., van der Mark P., et al, 2012. MrBayes 3. 2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol, 61(3): 539-542. DOI:10.1093/sysbio/sys029 |
Silvestro D., Michalak I., 2012. raxmlGUI: a graphical front-end for RAxML. Org. Divers. Evol, 12(4): 335-337. DOI:10.1007/s13127-011-0056-0 |
Spicer R.A., Harris N.B.W., Widdowson M., et al, 2003. Constant elevation of southern Tibet over the past 15 million years. Nature, 421: 622-624. DOI:10.1038/nature01356 |
Stamatakis A., 2014. RAxML Version 8: a tool for phylogenetic analysis and postanalysis of large phylogenies. Bioinformatics, 30(9): 1312-1313. DOI:10.1093/bioinformatics/btu033 |
Sun, H., 2013. Phytogeographical regions of China. In: Hong, D.Y., Blackmore, S. (Eds. ), Plants of China (A Companion to the Flora of China). Science Press, Beijing, pp. 176-204.
|
Sun H., Zhang J.W., Deng T., et al, 2017. Origins and evolution of plant diversity in the Hengduan Mountains, China. Plant Divers, 39(4): 161-166. DOI:10.1016/j.pld.2017.09.004 |
Sullivan J., Joyce P., 2005. Model selection in phylogenetics. Annu. Rev. Ecol. Evol. Syst, 36: 445-466. DOI:10.1146/annurev.ecolsys.36.102003.152633 |
Swofford, D.L., 2003. PAUP*: Phylogenetic Analysis Using Parsimony (And Other Methods), version 4.0a. Sinauer Associates Press, Sunderland.
|
Tu T.Y., Volis S., Dillon M.O., et al, 2010. Dispersals of hyoscyameae and mandragoreae (solanaceae) from the new world to eurasia in the early Miocene and their biogeographic diversification within eurasia. Mol. Phylogenet. Evol, 57(3): 1226-1237. DOI:10.1016/j.ympev.2010.09.007 |
Vassilczenko, I.T., 2000. Niedzwedzkia B. Fedtsch. In: Schischkin, B.K. (Ed. ), Flora of the USSR, Bignoniaceaee-Valerianaceae, Translated from Russian, vol. XXⅢ. Smithsonian Institute Libraries, Washington, pp. 5-7.
|
Wang, W.T., Pan, K.Y., Zhang, Z.Y., et al., 1990. Incarvillea Juss. In: Wang, W.T. (Ed. ), Flora Reipublicae Popularis Sinicae, vol. 69. Science Press, Beijing, pp. 34-49.
|
Wehr W.C., 1995. Early Tertiary flowers, fruits, and seeds of Washington State and adjacent areas. Wash. Geol, 23: 3-16. |
Wehr, W.C., Hopkins, D.Q., 1994. The Eocene Orchards and Gardens of Republic, vol. 22. Washington Geology, Washington, pp. 27-34.
|
Wehr, W.C., Manchester, S.R., 1996. Paleobotanical Significance of Eocene Flowers, Fruits, and Seeds from Republic, vol. 2. Washington Geology, Washington, pp. 25-27.
|
White, T.J., Bruns, T., Lee, S., et al., 1990. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis, M.A., Gelfand, D.H., Sninsky, J.J., White, T.J. (Eds. ), PCR Protocols: a Guide to Methods and Applications. Academic Press, California, pp. 315-322.
|
Wilf P., 1997. When are leaves good thermometers? A new case for leaf margin analysis. Paleobiology, 23(3): 373-390. DOI:10.1017/S0094837300019746 |
Wu, Z.Y., Wu, S.G., 1996. A proposal for a new floristic kingdom (realm)-the Asiatic kingdom, its delineation and characteristics. In: Zhang, A.L., Wu, S.G. (Eds. ), Floristics Characteristics and Diversity of East Asian Plants. China Higher Education, Beijing, pp. 3-42.
|
Xu B., Li Z.M., Sun H., 2014. Plant diversity and floristic characters of the alpine subnival belt flora in the Hengduan Mountains, SW China. J. Syst. Evol, 52(3): 271-279. DOI:10.1111/jse.12037 |
Xu T.T., Abbott R.J., Milne R.I., et al, 2010. Phylogeography and allopatric divergence of cypress species (Cupressus L. ) in the Qinghai-Tibetan Plateau and adjacent regions. BMC Evol. Biol, 10: 194. DOI:10.1186/1471-2148-10-194 |
Yu Y., Harris A.J., Blair C., et al, 2015. RASP (reconstruct ancestral state in phylogenies): a tool for historical biogeography. Mol. Phylogenet. Evol, 87: 46-49. DOI:10.1016/j.ympev.2015.03.008 |
Yue J.P., Sun H., Baum D.A., et al, 2009. Molecular phylogeny of Solms-laubachia(Brassicaceae) s. l., based on multiple nuclear and plastid DNA sequences, and its biogeographic implications. J. Syst. Evol, 47(5): 402-415. DOI:10.1111/j.1759-6831.2009.00041.x |
Zachos J., Pagani M., Sloan L., et al, 2001. Trends, rythms, and aberrations in global climate 65 Ma to present. Science, 292(5517): 686-693. DOI:10.1126/science.1059412 |
Zhang J.Q., Meng S.Y., Allen G.A., et al, 2014. Rapid radiation and dispersal out of the Qinghai-Tibetan Plateau of an alpine plant lineage Rhodiola (Crassulaceae). Mol. Phylogenet. Evol, 77: 147-158. DOI:10.1016/j.ympev.2014.04.013 |
Zhang Y.L., Li B.Y., Zheng D., 2002. A discussion on the boundary and area of the Tibetan Plateau in China. Geogr. Res, 21(1): 1-8. |
Zhang, Z.Y., Santisuk, T., 1998. Incarvillea Juss. In: Wu, Z.Y., Raven, P.H., Hong, D.Y. (Eds. ), Flora of China, vol. 18. Science Press & Missouri Botanical Garden Press, Beijing & St. louis, pp. 220-224.
|
Zhao Q.S., 1985. A study on the genus Incarvillea Juss. Of the transverse mountain range. Explor. Nature, 12: 170-174. |
Zhou Z., Hu J.J., Wen J., et al, 2019. Morphometric, phylogenetic and biogeographic analyses of Pyrularia (Santalales), a parasitic disjunct lineage between eastern Asia and eastern North America. Taxon, 68(1): 47-71. DOI:10.1002/tax.12021 |
Zhu H., Roos M.C., 2004. The tropical flora of southern China and its affinity to Indo-Malesian flora. Telopea, 10: 639-648. |
Zhu Y., Geng Y.P., Tersing T., et al, 2009. High genetic differentiation and low genetic diversity in Incarvillea younghusbandii, an endemic plant of QinghaiTibetan Plateau, revealed by AFLP markers. Biochem. Systemat. Ecol, 37(5): 589-596. DOI:10.1016/j.bse.2009.10.007 |



