b. Germplasm Bank of Wild Species, Kunming Institute of Botany, Chinese Academy of Sciences, Kunming, 650201, Yunnan, China;
c. University of Chinese Academy of Sciences, Beijing, 100049, China;
d. Department of Wetland, Southwest Forestry University, Kunming, 650224, China;
e. Center for Integrative Conservation, Xishuangbanna Tropical Botanical Garden, Chinese Academy of Sciences, Mengla, 666303, China;
f. Center of Conservation Biology, Core Botanical Gardens, Chinese Academy of Sciences, Mengla, 666303, China;
g. Department of Botany and Biodiversity Research, University of Vienna, Rennweg 14, A-1030, Vienna, Austria
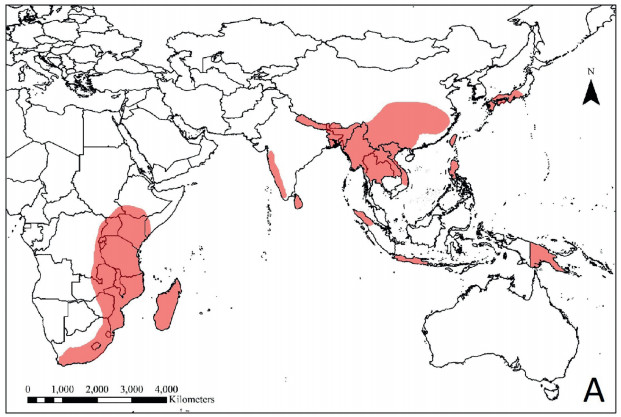
Dumasia DC. (Fabaceae: Papilionoideae: Phaseoleae) is widely distributed in tropical and subtropical regions of Asia, Africa, and in Papua New Guinea (Fig. 1), and its center of diversity is SW China (Lackey, 1981; Pradeep and Nayar, 1991). The genus was established by De Candolle, 1826, De Candolle, 1825 and 22 species names (including one hybrid) have been published for Dumasia up till now (Pan and Zhu, 2010). Sa and Gilbert (2010) recognized nine species distributed in China and indicated ca. 10 species occur globally. The most recent revision of the genus by Pan and Zhu (2010) recognized eight species, two subspecies and one variety of Dumasia worldwide, out of which seven species and one subspecies were distributed in China. Additionally, Pan et al. (unpublished results) reported the occurrence of Dumasia prazeri S.V. Pradeep & M.P. Nayar in China, which means all eight known species of Dumasia can be found in China (see Appendices S1 and S2). In their revision, Pan and Zhu (2010) also indicated that pubescence, stipules, leaflet shape, and pod shape are important diagnostic characters for keying out the species of Dumasia, while inflorescence length, flower length, flower dissections, and seed number are of little taxonomic significance. Nevertheless, though the composition of the genus is not disputed, the evolutionary relationships between species of Dumasia still need clarification. Furthermore, because there are few morphological differences between Dumasia species, plants are difficult to accurately identify. Accurate identification of specimens is not only critical for proper classification but misidentification may conceal cryptic species. Therefore, an accurate and rapid approach to identifying Dumasia species is needed.
|
| Fig. 1 The distribution of Dumasia around the world based on the distribution maps of each species of Dumasia provided by Pan and Zhu (2010). |
In the past few decades, molecular techniques have become increasingly popular for taxonomic studies (Doyle, 1992; Soltis et al., 2000; Moore et al., 2010). DNA barcoding is one of molecular methods of species identification that uses one or several short standard DNA regions (Kress et al., 2005; Hollingsworth et al., 2009). DNA barcoding is not only for species identification, but also for evolutionary, ecological, and conservation research (Hebert et al., 2003; Valentini et al., 2009; Li et al., 2011a; Liu et al., 2018; Leese et al., 2018). In addition, the use of DNA barcodes has also led to the discovery of many new and/or cryptic species (e. g., Hebert et al., 2004; Bączkiewicz et al., 2017; Tyagi et al., 2019). Accordingly, DNA barcoding has become an effective tool for uncovering hidden diversity and has enhanced our understanding of biodiversity (Gregory, 2005; Kress et al., 2005; Li et al., 2011b).
Cryptic species are defined as two or more distinct species that are classified (and hidden) under one species name, because they are superficially morphologically indistinguishable (Bickford et al., 2007; Struck et al., 2018). As a result of traditional taxonomy, which defines different species through macroscopic morphological differences, cryptic species are widely hidden in nature. The rise of DNA barcoding has provided a possible method for discovering cryptic species. So far, most cryptic diversity has been found in animals, especially invertebrates (e. g. Hebert et al., 2004; Witt et al., 2006; Pfenninger et al., 2007; Johnson et al., 2008; Lara et al., 2010; Brasier et al., 2016; Tyagi et al., 2017; Kanturski et al., 2018). In contrast, although roughly 6, 000 articles on cryptic species have been published (Struck et al., 2018), only a limited number have been found in seed plants. One significant example is in Taxus, where four cryptic species were revealed by DNA barcoding (Liu et al., 2011), and have subsequently been further supported by population genetics and morphological evidence (Liu et al., 2013, 2018; Möller et al., 2013). Carstens and Salter (2013) found that Sarracenia alata (Alph. Wood) Alph. Wood, a species distributed in SE USA, contains two cryptic lineages by using several modern methods (Gaussian clustering, Structurama, BPP and spedeSTEM). In the Fabaceae, Acacia s.l. contains many species which are difficult to define by macroscopic morphology, but Newmaster and Ragupathy (2009) showed that DNA barcoding can help identify cryptic species.
In this study, we used six DNA regions, psbB-psbF and five previously proposed barcodes (ITS, trnH-psbA, matK, rbcL, trnL-trnF) (Kress et al., 2005; Kress and Erickson, 2007; Taberlet et al., 2007; Lahaye et al., 2008; Hollingsworth et al., 2009, 2011; Li et al., 2011b), as markers to differentiate species of Dumasia. Our objectives were (1) to clarify the phylogenetic relationships among Dumasia species using nuclear and cpDNA sequences, (2) to test the utility of DNA barcoding in Dumasia, and (3) to validate the previous taxonomic treatments of Dumasia.
2. Materials and methods 2.1. Sampling strategyA total of 61 accessions of all eight currently recognized species of Dumasia (including two individuals of nominal species Dumasia nitida Chun ex Y.T. Wei & S.K. Lee, which was synonymized with Dumasia truncata Siebold & Zucc. by Pan and Zhu (2010)) were sampled, along with two individuals of Toxicopueraria peduncularis (Benth.) A. N. Egan & B. Pan bis as an outgroup (Appendix S1). Fresh leaves were immediately stored in silica gel and transported back to the laboratory for DNA extraction. Voucher specimens of the collected taxa were deposited in the herbaria HITBC, PE, and SWFC. The latitude, longitude, and altitude of each accession sampled were recorded using an Extrex GIS monitor (Garmin, Taiwan, China).
2.2. DNA isolation, PCR amplification, and sequencingGenomic DNA was isolated using the Plant Genomic DNA Kit (TIANGEN Biotech, Beijing, China), following the manufacturer's instructions. The DNA samples were stored at -20 ℃ prior to amplification. Polymerase chain reaction (PCR) was carried out in a 20 μL reaction volume containing 2.5 μL of 10 × buffer with 2 mM MgCl2, 1 U Taq DNA polymerase, 1 μL of dNTP (0.125 mM), 1 μL of each primer (5 pM), and 30–50 ng total DNA. Nuclease-free water was added to complete the final volume. The optimal PCR conditions and primer information are displayed in Appendix S3. We visualized PCR products (2 μL) on 0.8% agarose gels by electrophoresis. PCR products were purified using the BioMed multifunctional DNA fragment purification recovery kits (Beijing, China), and then sequenced with the same primers used for PCR amplifications in an ABI 3730 automated sequencer (Applied Biosystems, Carlsbad, California, USA).
2.3. Sequence alignment and data analysisThe sequences were aligned using MUSCLE (Edgar, 2004) in MEGA 7.0 (Kumar et al., 2016) and further checked manually. We used both single loci and all possible combinations of the six loci for the DNA barcoding survey. The intra- and interspecific divergences were calculated based on the Kimura-2-parameter (K2P) model in MEGA 7.0 (Kumar et al., 2016). To detect the presence of a barcoding gap for each species, the minimum interspecific distances and maximum intraspecific distances were compared in order (Meyer and Paulay, 2005; Zhang et al., 2015). To assess the accuracy of barcodes for species assignment, the functions of the 'best match' and the 'best close match' method in the program TaxonDNA (Meier et al., 2006) were used. The Wilcoxon signed-ranks test is widely used for the examination of the barcoding gap (Kress and Erickson, 2007; Lahaye et al., 2008; Zhang et al., 2015; Girma et al., 2016). In this study, we used the Wilcoxon signed-ranks test based on the K2P model and the pairwise distances (p-distances) to assess the differences between intraspecific and interspecific divergences within each pair of barcodes in PASW Statistics 18.0 (formerly SPSS Statistics).
Phylogenetic analyses with combinations of ITS and the five cpDNA markers were performed with standard Bayesian inference (BI), maximum-parsimony (MP), maximum-likelihood (ML), and neighbor-joining (NJ) methods. The optimal fitting model was determined by MODELTEST v.3.7 (Posada and Crandall, 1998) with the Akaike Information Criterion (AIC) (Posada and Buckley, 2004). The BI analysis was performed with MrBayes v.3.2 (Ronquist et al., 2012) with four Markov Chain Monte Carlo (MCMC) runs using a random starting tree, an invgamma rate model with six discrete categories and ten million generations, with a sampling frequency of one every 1, 000 generations and 25% of the trees discarded as burn-in. Stationarity was considered to be reached when the average standard deviations of split frequencies were below 0.01. The ML analysis was performed with command line RAxML v.7.2.8 (Stamatakis, 2006) in Linux OS, including tree robustness assessment using 100 replicates of rapid bootstrap (the "-f a" option) with the GTR + G + I substitution model to assess branch support. The NJ analysis was performed with MEGA v.7.0 using the K2P model and the node support was assessed with 1, 000 bootstrap replicates. The MP analysis was also performed with MEGA v.7.0 and the node support was assessed with 500 bootstrap replicates. In addition, phylogenetic analyses were performed separately on the ITS and plastid DNA matrices using Bayesian inference and maximum-likelihood models.
3. Results 3.1. Barcode universality and sequence characteristicsWe obtained sequences from all accessions of the eight Dumasia species and Toxicopueraria peduncularis (as outgroup) with 100% PCR success and 100% sequencing success. Only universal primers were used. The ITS matrix contained 744 bp and 10 indels 1–10 bp long; the distribution of 156 bp parsimony informative sites and 160 bp variable sites was intensive and dense across the matrix. The trnH-psbA matrix contained 391 bp and 2 indels 18–34 bp long; the distribution of 8 parsimony informative sites and 8 variable sites was dispersive and sparse across the matrix. For the matK matrix, the length of aligned sequences was 870 bp; the distribution of 6 parsimony informative sites and 8 variable sites was dispersive and sparse across the matrix, and there was 1 indel 18 bp long. For the rbcL matrix, the aligned sequence length was 575 bp; the distribution of 5 parsimony informative sites and 5 variable sites was dispersive and sparse across the matrix, without indels. For trnL-trnF matrix, aligned sequence length was 721 bp, with 3 indels 1–21 bp long; the distribution of 4 parsimony informative sites and 4 variable sites was dispersive and sparse across the matrix. As for psbB-psbF matrix, it contained 776 bp and 1 indel 1 bp long; the distribution of 6 parsimony informative sites and 7 variable sites was dispersive and sparse across the matrix. The average interspecific distances as determined by each of the six DNA regions were 7.33%, 0.73%, 0.26%, 0.30%, 0.15%, and 0.22%, respectively, while the average intraspecific distances were 0.73%, 0.21%, 0.01%, 0.06%, 0.01%, and 0.04%. All sequence characteristics of the six DNA regions of Dumasia mentioned above are shown in Table 1.
| ITS | matK | psbA-trnH | psbB-psbF | rbcL | trnL-trnF | |
| Universality of primers | Yes | Yes | Yes | Yes | Yes | Yes |
| Percentage PCR success | 100 | 100 | 100 | 100 | 100 | 100 |
| Percentage sequencing success | 100 | 100 | 100 | 100 | 100 | 100 |
| No. of species (individuals) | 8 (59) | 8 (59) | 8 (59) | 8 (59) | 8 (59) | 8 (59) |
| Aligned sequence length (bp) | 744 | 870 | 391 | 776 | 575 | 721 |
| No. of parsimony-informative sites | 156 | 6 | 8 | 6 | 5 | 4 |
| No. of variable sites | 160 | 8 | 8 | 7 | 5 | 4 |
| No. of indels (length range) | 10 (1–10) | 1 (18) | 2 (18–34) | 1 (1) | 0 | 3 (1–21) |
| Average interspecific distance (range) (%) | 7.33 (4.04–11.64) | 0.26 (0–0.59) | 0.73 (0.30–1.79) | 0.22 (0–0.42) | 0.30 (0–0.70) | 0.15 (0–0.43) |
| Average intraspecific distance (range) (%) | 0.73 (0–2.46) | 0.01 (0–0.06) | 0.21 (0–0.89) | 0.04 (0–0.26) | 0.06 (0–0.26) | 0.01 (0–0.09) |
Most DNA markers or their combinations showed relatively clear barcoding gaps between intraspecific and interspecific genetic variation, such as the combination of ITS and the 5 cpDNA markers (Fig. 2A). For single barcodes, ITS showed the most obvious barcoding gap between intraspecific and interspecific genetic distance (Fig. 2B).

|
| Fig. 2 Relative distribution of inter- and intraspecific distances of the combination of six DNA barcoding markers (A) and ITS sequences of Dumasia (B). |
The P-distance-based Wilcoxon signed-ranks test reflects divergences between different barcoding markers more clearly than the K2P-based Wilcoxon signed-ranks test, regardless of whether examining interspecific variation or intraspecific divergence (Table 2, Table 3). For interspecific divergence, ITS showed the largest divergence among the six barcoding markers, while trnL-trnF sequence showed the smallest divergence. The order from large to small was ITS > psbA-trnH > rbcL > matK > psbB-psbF > trnL-trnF. For intraspecific divergence, ITS again showed the largest variation, whereas the differences among matK, psbB-psbF, and trnL-trnF were not significant. The order of intraspecific divergence from large to small was ITS > psbA-trnH > rbcL > matK = psbB-psbF = trnL-trnF.
| W+ | W- | Relative ranks | N | p-value ≤ | Result | |
| W+ | W- | |||||
| interspecific distance | ||||||
| ITS | matK | 666 | 0 | 36 | 1.74E-07 | ITS > matK |
| ITS | psbA-trnH | 665 | 1 | 36 | 1.90E-07 | ITS > psbA-trnH |
| ITS | psbB-psbF | 666 | 0 | 36 | 1.74E-07 | ITS > psbB-psbF |
| ITS | rbcL | 666 | 0 | 36 | 1.75E-07 | ITS > rbcL |
| ITS | trnL-trnF | 666 | 0 | 36 | 1.73E-07 | ITS > trnL-trnF |
| matK | psbA-trnH | 0 | 630 | 36 | 2.53E-07 | matK < psbA-trnH |
| matK | psbB-psbF | 274.5 | 160.5 | 36 | 0.2093 | non-significant |
| matK | rbcL | 239 | 226 | 36 | 0.8995 | non-significant |
| matK | trnL-trnF | 540 | 21 | 36 | 3.13E-06 | matK > trnL-trnF |
| psbA-trnH | psbB-psbF | 630 | 0 | 36 | 2.49E-07 | psbA-trnH > psbB-psbF |
| psbA-trnH | rbcL | 594 | 1 | 36 | 4.03E-07 | psbA-trnH > rbcL |
| psbA-trnH | trnL-trnF | 666 | 0 | 36 | 1.70E-07 | psbA-trnH > trnL-trnF |
| psbB-psbF | rbcL | 118 | 158 | 36 | 0.5477 | non-significant |
| psbB-psbF | trnL-trnF | 515.5 | 12.5 | 36 | 1.55E-06 | psbB-psbF > trnL-trnF |
| rbcL | trnL-trnF | 478.5 | 17.5 | 36 | 5.36E-06 | rbcL > trnL-trnF |
| intraspecific distance | ||||||
| ITS | matK | 20 | 1 | 9 | 0.0592 | non-significant |
| ITS | psbA-trnH | 18 | 3 | 9 | 0.1422 | non-significant |
| ITS | psbB-psbF | 20 | 1 | 9 | 0.0592 | non-significant |
| ITS | rbcL | 20 | 1 | 9 | 0.0592 | non-significant |
| ITS | trnL-trnF | 15 | 0 | 9 | 0.0591 | non-significant |
| matK | psbA-trnH | 1 | 9 | 9 | 0.2012 | non-significant |
| matK | psbB-psbF | 6 | 4 | 9 | 0.8551 | non-significant |
| matK | rbcL | 3 | 7 | 9 | 0.5839 | non-significant |
| matK | trnL-trnF | 6 | 0 | 9 | 0.1814 | non-significant |
| psbA-trnH | psbB-psbF | 6 | 0 | 9 | 0.1814 | non-significant |
| psbA-trnH | rbcL | 6 | 0 | 9 | 0.1814 | non-significant |
| psbA-trnH | trnL-trnF | 6 | 0 | 9 | 0.1814 | non-significant |
| psbB-psbF | rbcL | 2 | 4 | 9 | 0.7893 | non-significant |
| psbB-psbF | trnL-trnF | 6 | 0 | 9 | 0.1814 | non-significant |
| rbcL | trnL-trnF | 6 | 0 | 9 | 0.1814 | non-significant |
| The symbols "W+"and "W-" represent the sums of all positive and negative values in the signed-rank column, respectively. Symbol " > " is used if the interspecific or intraspecific divergence for one barcoding marker significantly exceeds that of another barcoding marker. | ||||||
| W+ | W- | Relative ranks | N | p-value ≤ | Result | |
| W+ | W- | |||||
| interspecific distance | ||||||
| ITS | matK | 1006071 | 0 | 1418 | 2.20E-16 | ITS > matK |
| ITS | psbA-trnH | 1006050 | 21 | 1418 | 2.20E-16 | ITS > psbA-trnH |
| ITS | pabB-psbF | 1006071 | 0 | 1418 | 2.20E-16 | ITS > psbB-psbF |
| ITS | rbcL | 1006071 | 0 | 1418 | 2.20E-16 | ITS > rbcL |
| ITS | trnL-trnF | 1006071 | 0 | 1418 | 2.20E-16 | ITS > trnL-trnF |
| matK | psbA-trnH | 79101 | 814015 | 1418 | 2.20E-16 | matK < psbA-trnH |
| matK | pabB-psbF | 611438 | 259102 | 1418 | 2.20E-16 | matK > psbB-psbF |
| matK | rbcL | 344517 | 510261 | 1418 | 1.21E-09 | matK < rbcL |
| matK | trnL-trnF | 812996 | 28757 | 1418 | 2.20E-16 | matK > trnL-trnF |
| psbA-trnH | psbB-psbF | 769892 | 47389 | 1418 | 2.20E-16 | psbA-trnH > psbB-psbF |
| psbA-trnH | rbcL | 768844 | 99059 | 1418 | 2.20E-16 | psbA-trnH > rbcL |
| psbA-trnH | trnL-trnF | 849961 | 3510 | 1418 | 2.20E-16 | psbA-trnH > trnL-trnF |
| psbB-psbF | rbcL | 157341 | 692215 | 1418 | 2.20E-16 | psbB-psbF < rbcL |
| psbB-psbF | trnL-trnF | 728634 | 49494 | 1418 | 2.20E-16 | psbB-psbF > trnL-trnF |
| rbcL | trnL-trnF | 761261 | 10642 | 1418 | 2.20E-16 | rbcL > trnL-trnF |
| intraspecific distance | ||||||
| ITS | matK | 50389 | 3896 | 412 | 2.20E-16 | ITS > matK |
| ITS | psbA-trnH | 43077 | 10879 | 412 | 2.20E-16 | ITS > psbA-trnH |
| ITS | psbB-psbF | 46377 | 2764 | 412 | 2.20E-16 | ITS > psbB-psbF |
| ITS | rbcL | 40557 | 1638 | 412 | 2.20E-16 | ITS > rbcL |
| ITS | trnL-trnF | 38397 | 106 | 412 | 2.20E-16 | ITS > trnL-trnF |
| matK | psbA-trnH | 4560 | 32841 | 412 | 2.20E-16 | matK < psbA-trnH |
| matK | psbB-psbF | 20725 | 16403 | 412 | 0.09353 | non-significant |
| matK | rbcL | 8364 | 22761 | 412 | 2.08E-10 | matK < rbcL |
| matK | trnL-trnF | 19315 | 10088 | 412 | 2.02E-05 | matK > trnL |
| psbA-trnH | psbB-psbF | 29386 | 2745 | 412 | 2.20E-16 | psbA-trnH > psbB-psbF |
| psbA-trnH | rbcL | 29470 | 5246 | 412 | 2.20E-16 | psbA-trnH > rbcL |
| psbA-trnH | trnL-trnF | 31100 | 1540 | 412 | 2.20E-16 | psbA-trnH > trnL-trnF |
| psbB-psbF | rbcL | 4307 | 28333 | 412 | 2.20E-16 | psbB-psbF < rbcL |
| psbB-psbF | trnL-trnF | 16065 | 12138 | 412 | 0.06128 | non-significant |
| rbcL | trnL-trnF | 15931 | 0 | 412 | 2.20E-16 | rbcL > trnL-trnF |
| The symbols "W+" and "W-" represent the sums of the positive and negative values in the signed-rank column, respectively. Symbol " > " is used if the interspecific or intraspecific divergence for one barcoding marker significantly exceeds that of another barcoding marker. | ||||||
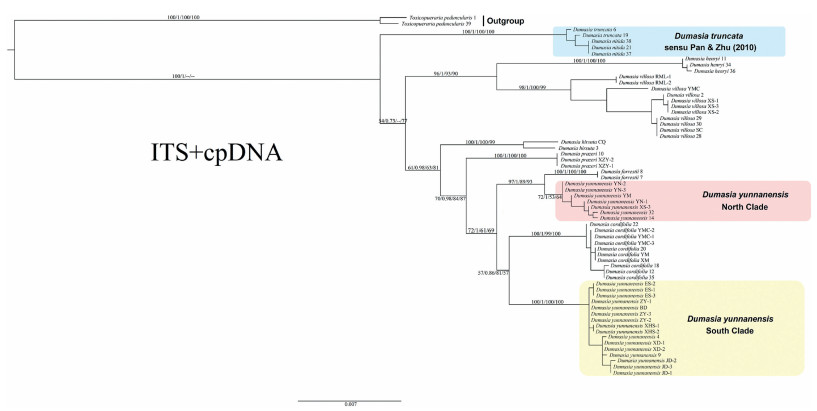
The NJ, ML, Bayesian, and MP analyses of the combined data set of ITS and five chloroplast markers generated fairly similar topologies (Figs. 3-5). With the exception of Dumasia yunnanensis Y. T. Wei & S. K. Lee, all species were monophyletic, with strong support except in the poorly resolved cpDNA tree. D. yunnanensis was separated into two non-sister clades in all trees, with one clade sister to Dumasia forrestii Diels and the other sister to Dumasia cordifolia Benth. ex Baker.
|
| Fig. 3 Phylogenetic relationships among 59 individuals from eight species of Dumasia and two individuals of Toxicopueraria peduncularis based on the combination of ITS and five cpDNA markers. The tree was constructed using the Maximum Likelihood method. Numbers on branches are bootstrap percentages (BP) and posterior probabilities (PP) from Maximum Likelihood (ML), Bayesian analysis, Neighbor-joining analysis, and Maximum-parsimony analyses, respectively. A dash (-) indicates the topologies generated from the other three methods are different from the ML method. |
|
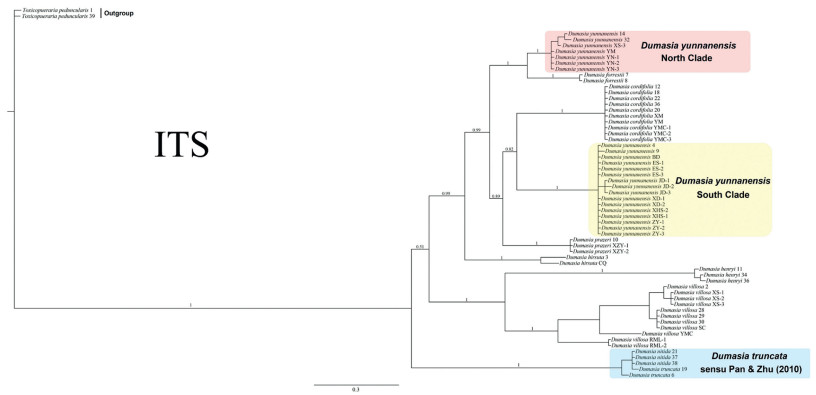
| Fig. 4 Phylogenetic tree for Dumasia from Bayesian Inference based on ITS. Numbers on branches indicate the posterior probabilities (PP) from Bayesian analysis. |

|
| Fig. 5 Phylogenetic tree for Dumasia obtained from Bayesian Inference (BI) based on the combination of five cpDNA markers. Numbers on branches indicate the posterior probabilities (PP) of the BI tree for cpDNA. |
The ITS tree (Fig. 4) had a different topology from the cpDNA tree (Fig. 5). In the ITS tree, D. truncata (including D. nitida), Dumasia henryi (Hemsl. ex F. B. Forbes & Hemsl.) R. Sa & M. G. Gilbert + Dumasia villosa DC. (sister to each other), and Dumasia hirsuta Craib form a paraphyletic grade successively, while D. hirsuta is sister to all remaining taxa. The remaining taxa form three major clades: D. yunnanensis (I) and its sister D. forrestii form the first clade, which is sister to the other two clades; D. prazeri forms the second clade, which is sister to the third clade, D. yunnanensis (II) and its sister group D. cordifolia. However, in the plastid-only tree, the genus can be separated into two sister clades. One clade consists of D. villosa, D. prazeri, D. hirsuta, and D. henryi, with the clade of D. henryi + D. hirsuta, with low posterior probabilities, sister to the clade of D. villosa and D. prazeri, which are sister to each other. The other clade includes the remaining species, with D. truncata (including D. nitida) basal, D. yunnanensis (I) allied with D. forrestii, and D. yunnanensis (II) allied with D. cordifolia, with these two clades sister to each other, although the whole four species are not resolved as monophyletic. The tree based on the combined ITS plus cpDNA data had a topology almost congruent with the ITS tree (Fig. 3).
3.4. Rates of identificationThe rates of sample identification with each DNA barcode and their combinations are shown in Appendix S4. ITS and any combination that included ITS had the highest success rate for correct identification of species (> 96.6%). Least success was obtained with trnL-trnF (5%).
4. Discussion 4.1. DNA barcoding provides a new method to identify Dumasia speciesA suitable DNA barcode must show high rates of universal primer amplification and sequencing, as well as a strong ability to identify and discriminate species (Kress et al., 2005; Kress and Erickson, 2007). The six DNA fragments used in this study all had 100% success rates for PCR amplification and sequencing (Table 1), but ITS had a much higher overall species discrimination than the others, as also reported in a previous study (Li et al., 2011b). ITS also showed the best barcoding gap, species resolution, and identification efficiency (Fig. 2B, Table 2, Table 3, Appendix S4). Furthermore, the combination of ITS and any one of the five plastid DNA markers used in this study also achieve very high species resolution. The high-resolution ability of ITS may be attributed to its high evolution rate, leading to genetic changes that can distinguish closely related species in the same genus (Kress et al., 2005; Liu et al., 2011). The other four barcodes (matK, rbcL, psbA-trnH and trnL-trnF) used in this study have all been proposed as core or supplementary regions for plant barcoding (Kress et al., 2005; CBOL Plant Working Group, 2009; Chen et al., 2010; Hollingsworth et al., 2011), but together with the additional plastid region psbB-psbF exhibited low species-level resolution in our study. The low resolution of plastid regions at the species level has also been reported in other plants previously (Li et al., 2016; Liu et al., 2017) and may reflect the lower substitution rates found in plastid genomes compared to nuclear genomes. Previous studies have shown that DNA barcode combinations can improve species discrimination (CBOL Plant Working Group, 2009; Li et al., 2011a); however, in this study ITS provided high species-level resolution whether it was used alone or in combination with other barcoding regions. Therefore, we suggest that ITS should be used alone as a barcode to identify Dumasia species.
4.2. Molecular phylogeny can provide evidence for taxonomic treatmentWei and Lee (1985) first described D. nitida as a new species, similar to D. truncata. but differing in its 5–13 cm long inflorescence, loose arrangement of flowers on the rachis, and pods with only 1–2 seeds. Pan and Zhu (2010) synonymized D. nitida with D. truncata based on the examination of a large number of specimens. The present study shows that D. truncata and D. nitida cannot be separated by the barcodes used (shown as the sky-blue box in Figs. 3 and 4), supporting the treatment of D. nitida as a synonym of D. truncata.
Forbes and Hemsley (1886–1888) first described Rhynchosia henryi Hemsl. and noted that this species is distinct in its tubular truncate calyx, more or less fissured on the vexillary side. Merrill (1910) synonymized it with D. villosa. R. henryi differs from D. villosa only in the shape of leaflets, oblong vs. ovate to broad ovate, and the wing petals of R. henryi are also larger than in D. villosa. Wei and Lee (1985) described Dumasia oblongifoliolata F. T. Wang & Tang ex Y. T. Wei & S. K. Lee, which should be conspecific with R. henryi. Sa and Gilbert (2010) published the new combination D. henryi and cited D. oblongifoliolata as a synonym. Pan and Zhu (2010) followed this treatment. In this study, the phylogenetic tree shows that D. henryi and D. villosa each form separate clades, with high bootstrap percentages (BP) and posterior probabilities (PP), which are sister to each other (Fig. 3), strongly supporting the previous treatments (Sa and Gilbert, 2010; Pan and Zhu, 2010).
Regrettably, we did not sample any of the infraspecific taxa accepted by Pan and Zhu (2010) or any material from outside mainland China, and, therefore, we cannot resolve the systematic position of these taxa.
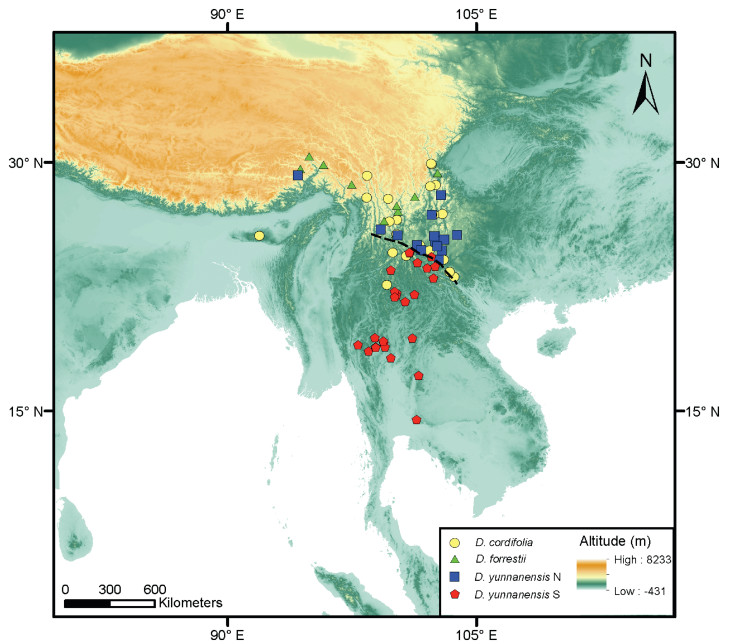
4.3. DNA barcoding reveals cryptic lineage in Dumasia yunnanensisDetecting cryptic species is the one of most appealing applications of DNA barcoding (Hebert et al., 2004; Gao et al., 2017). In our phylogenetic trees, D. yunnanensis is clearly separated into two groups (see Results and Figs. 3-5): one (I, in red box, North Clade) allied to D. forrestii and another (II, in yellow box, South Clade) allied to D. cordifolia. These two groups are morphologically indistinguishable and have been regarded as conspecific in past taxonomic treatments (Sa and Gilbert, 2010; Pan and Zhu, 2010). However, they have distinct geographical distributions: (I) is distributed north of the range of (II) (see Fig. 6). This points to the existence of cryptic species in D. yunnanensis. Previous studies have shown that D. cordifolia can be distinguished from other Dumasia taxa by its cordiform leaflets on upper leaves (vs. never cordiform in D. yunnanensis) while D. forrestii can be distinguished from D. yunnanensis by its round leaflets and tetragonal stems (vs. ovate leaflets and terete stem in D. yunnanensis) (Pan and Zhu, 2010) (Fig. 7).
|
| Fig. 6 The distribution of the collection locations of herbarium specimens of four taxa mainly discussed in this paper, the dotted line separates the distributions of two clades of D. yunnanensis. The map was prepared by Dr. Shu-feng Li based on collection locations of herbarium specimens provided by Pan and Zhu (2010) and Meeboonya et al. (2019). |
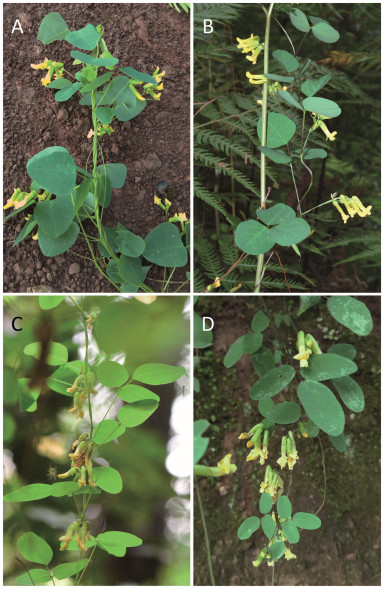
|
| Fig. 7 The four taxa of Dumasia most discussed in this paper: (A) D. cordifolia; (B) D. forrestii; (C) D. yunnanensis north clade, and (D) D. yunnanensis south clade. A was photographed by Bo Pan, B by Dr. Bing Liu, C by Dr. Ren-bin Zhu, and D by Mr. Yi Fu. |
Cryptic species could result from recent divergence, parallelism, convergence, or stasis (Struck et al., 2018). When cryptic species arise as a result of recent evolutionary divergence and stasis, these species should be sister taxa or members of a species complex. In this study, the two hypothesized cryptic species are not sister taxa, and, in both the ITS and cpDNA phylogenetic trees, they group with D. cordifolia and D. forrestii, respectively (Figs. 2, 4 and 5). Therefore, we speculate that the most likely causes of cryptic species in D. yunnanensis are parallelism or convergence.
The samples of D. yunnanensis also clustered in two clades in the cpDNA tree (Fig. 5). However, in contrast to the ITS tree, they were not clearly differentiated from the closely related species, D. cordifolia and D. forestii, respectively, in contrast to the ITS tree (Fig. 4). This incongruence may result from either hybridization and introgression or incomplete lineage sorting, as has been found in many previous studies using cpDNA (Li et al., 2011b). The specific causes of this phenomenon and the details of the cryptic differentiation need further exploration using multiple, genome-wide, highly polymorphic markers. Only four populations from the Northern group were sampled in the current study, so more comprehensive sampling of populations within the range of the D. yunnanensis is also needed, particularly at the peripheral extent of its range.
5. ConclusionsThis study provides comparative assessments of six candidate barcoding loci and their combinations for resolving species of Dumasia (Fabaceae: Phaseoleae). Our results show that ITS is the best barcoding sequence for Dumasia plants. ITS has the highest discriminatory power, and can distinguish between all Dumasia species when used alone or in combinations with any cpDNA barcodes tested. Our phylogenetic analysis of Dumasia using barcode sequences confirmed the most recent taxonomic treatment, except that it revealed two evolutionarily distinct lineages in D. yunnanensis, which have allopatric distributions and appear to be cryptic species. Together with previous cases (e. g., Liu et al., 2011; Carstens and Salter, 2013), the discovery of putative new species in this small genus suggests that our current knowledge of species diversity is not yet complete, and it is feasible to use molecular tools to find them.
Author contributionsBin Tian and Bo Pan designated the study and managed the project. Bo Pan and Bin Tian collected samplings in which Bo Pan completed the species identification. Bin Tian prepared DNA samples and performed sequencing. Kai-wen Jiang, Rong Zhang and Bin Tian performed the DNA barcoding and molecular phylogenetic analyses. Rong Zhang and Zhong-fu Zhang performed Wilcoxon signed-rank tests. Kai-wen Jiang wrote the manuscript. All authors read and approved the final manuscript.
Declaration of competing interestThere is no known Conflict of Interest in this paper.
AcknowledgementsWe thank Dr. Zhi-qiang Lu and Mr. Yi Fu for help during the field survey. We are grateful to Dr. Ovidiu Paun for very helpful comments on earlier drafts of this manuscript. We thank Dr. Shu-feng Li for the distributional map, as well as Dr. Bing Liu, Dr. Ren-bin Zhu, and Mr. Yi Fu for their photos of some Dumasia species. The first author thanks Dr. Wen-bin Yu, Dr. Pei-liang Liu, Dr. Xue-li Zhao, and Dr. Zhu-qiu Song for their help during the writing process. Additional thanks go to Dr. Richard T. Corlett, Raymond Porter and Mr Yuan-qiong Zhang for polishing this work. The authors would also like to express gratitude to two anonymous reviewers for their valuable comments on the manuscript. This work was financially supported by the Second Tibetan Plateau Scientific Expedition and Research (STEP) program (2019QZKK0502), the National Natural Science Foundation of China (NSFC 41861008) and the 135 Karst 'breakthrough' project Grant 2017XTBG-T03.
Appendix A. Supplementary dataSupplementary data to this article can be found online at https://doi.org/10.1016/j.pld.2020.07.007.
Bączkiewicz A., Szczecińska M., Sawicki J., et al, 2017. DNA barcoding, ecology and geography of the cryptic species of Aneura pinguis and their relationships with Aneura maxima and Aneura mirabilis (Metzgeriales, Marchantiophyta). PloS One, 12: e0188837. DOI:10.1371/journal.pone.0188837 |
Bickford D., Lohman D.J., Sodhi N.S., et al, 2007. Cryptic species as a window on diversity and conversation. Trends Ecol. Evol, 22: 148-155. DOI:10.1016/j.tree.2006.11.004 |
Brasier M.J., Wiklund H., Neal L., et al, 2016. DNA barcoding uncovers cryptic diversity in 50% of deep-sea Antarctic polychaetes. R. Soc. Open Sci, 3: 160432. DOI:10.1098/rsos.160432 |
Carstens B.C., Salter J.D., 2013. The carnivorous plant described as Sarracenia alata contains two cryptic species. Biol. J. Linn. Soc, 109: 737-746. DOI:10.1111/bij.12093 |
CBOL Plant Working Group, 2009. A DNA barcode for land plants. Proc. Natl. Acad.Sci. Unit. States Am, 106: 12794-12797. DOI:10.1073/pnas.0905845106 |
Chen S., Yao H., Han J., et al, 2010. Validation of the ITS2 region as a novel DNA barcode for identifying medicinal plant species. PloS One, 5: e8613. DOI:10.1371/journal.pone.0008613 |
De Candolle A.P., 1825. Dumasia DC. Ann. Sci. Nat., Zool, 4: 96-97. |
De Candolle, A.P., 1826. Dumasia DC. Mémoires sur la Famille des Légumineuses. A. Belin, Paris, pp. 255-257.
|
Doyle J.J., 1992. Gene trees and species trees: molecular systematics as onecharacter taxonomy. Syst. Bot, 17: 144-163. DOI:10.2307/2419070 |
Edgar R.C., 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res, 32: 1792-1797. DOI:10.1093/nar/gkh340 |
Forbes, F.B., Hemsley, W.B., 1886-1888. An enumeration of all the plants known from China Proper, Formosa, Hainan, Corea the Luchu Archipelago and the Island of Hongkong, together with their distribution and synonym. J. Linn. Soc.Bot. 23, 1-489.
|
Gao L.-M., Li Y., Phan L.-K., et al, 2017. DNA barcoding of East Asian Amentotaxus(Taxaceae): potential new species and implications for conservation. J. Systemat. Evol, 55: 16-24. DOI:10.1111/jse.12207 |
Gregory T.R., 2005. DNA barcoding does not compete with taxonomy. Nature, 434: 1067. |
Girma G., Spillane C., Gedil M., 2016. DNA barcoding of the main cultivated yams and selected wild species in the genus Dioscorea. J. Systemat. Evol, 54: 228-237. |
Hebert P.D.N., Cywinska A., Ball S.L., et al, 2003. Biological identifications through DNA barcodes. Proc. R. Soc. B, 270: 313-321. DOI:10.1098/rspb.2002.2218 |
Hebert P.D.N., Penton E.H., Burns J.M., et al, 2004. Ten species in one: DNA barcoding reveals cryptic species in the neotropical skipper butterfly Astraptes fulgerator. Proc. Natl. Acad. Sci. U.S.A, 101: 14812-14817. DOI:10.1073/pnas.0406166101 |
Hollingsworth P.M., Forrest L.L., Spouge J.L., et al, 2009. A DNA barcode for land plants. Proc. Natl. Acad. Sci. U.S.A, 106: 12794-12797. DOI:10.1073/pnas.0905845106 |
Hollingsworth P.M., Graham S.W., Little D.P., 2011. Choosing and using a plant DNA barcode. PloS One, 6: e19254. DOI:10.1371/journal.pone.0019254 |
Johnson S.B., Warén A., Vrijenhoek R.C., 2008. DNA barcoding of Lepetodrilus limpets reveals cryptic species. J. Shellfish Res, 27: 43-51. DOI:10.2983/0730-8000(2008)27[43:DBOLLR]2.0.CO;2 |
Kanturski M., Lee Y., Choi J., et al, 2018. DNA barcoding and a precise morphological comparison revealed a cryptic species in the Nippolachnus piri complex(Hemiptera: aphididae: Lachninae). Sci. Rep, 8: 8998. DOI:10.1038/s41598-018-27218-2 |
Kress W.J., Wurdack K.J., Zimmer E.A., et al, 2005. Use of DNA barcodes to identify flowering plants. Proc. Natl. Acad. Sci. U.S.A, 102: 8369-8374. DOI:10.1073/pnas.0503123102 |
Kress W.J., Erickson D.L., 2007. A two locus global DNA barcode for land plants: the coding rbcL gene complements the non-coding trnH-psbA spacer region. PloS One, 2: e508. DOI:10.1371/journal.pone.0000508 |
Kumar S., Stecher G., Tamura K., 2016. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol, 33: 1870-1874. DOI:10.1093/molbev/msw054 |
Lahaye R., van der Bank M., Bogarin D., et al, 2008. DNA barcoding the floras of biodiversity hotspots. Proc. Natl. Acad. Sci. U.S.A, 105: 2923-2928. DOI:10.1073/pnas.0709936105 |
Lackey, J.A., 1981. Tribe 10. Phaseoleae DC. In: Polhill, R.M., Raven, P.H. (Eds.), Advances in Legume Systematics, Part 1. Kew Publisher, Royal Botanical Gardens, Kew, pp. 301-327.
|
Lara A., de León J.L.P., Rodríguez R., et al, 2010. DNA barcoding of Cuban fresh-water fishes: evidence for cryptic species and taxonomic conflicts. Mol. Ecol.Resour, 10: 421-430. DOI:10.1111/j.1755-0998.2009.02785.x |
Leese F., Bouchez A., Abarenkov K., et al, 2018. Why we need sustainable networks bridging countries, disciplines, cultures and generations for aquatic Biomonitoring 2. 0: a perspective derived from the DNAqua-Net COST action. Adv.Ecol. Res, 58: 63-99. DOI:10.1016/bs.aecr.2018.01.001 |
Li D.-Z., Liu J.-Q., Chen Z.-D., et al, 2011a. Plant DNA barcoding in China. J. Systemat. Evol, 49: 165-168. DOI:10.1111/j.1759-6831.2011.00137.x |
Li D.-Z., Gao L.-M., Li H.-T., et al, 2011b. Comparative analysis of a large dataset indicates that internal transcribed spacer (ITS) should be incorporated into the core barcode for seed plants. Proc. Natl. Acad. Sci. Unit. States Am, 108: 19641-19646. DOI:10.1073/pnas.1104551108 |
Li Y.-L., Tong Y., Xing F.-W., 2016. DNA barcoding evaluation and its taxonomic implications in the recently evolved genus Oberonia Lindl. (Orchidaceae) in China. Front. Plant Sci, 7: 1791. |
Liu J., Möller M., Gao L.-M., et al, 2011. DNA barcoding for the discrimination of Eurasian yews (Taxus L., Taxaceae) and the discovery of cryptic species. Mol.Ecol. Res, 11: 89-100. DOI:10.1111/j.1755-0998.2010.02907.x |
Liu J., Milne R.I., Moller M., et al, 2018. Integrating a comprehensive DNA barcode reference library with a global map of yews (Taxus L.) for forensic identification. Mol. Ecol. Res, 18: 1115-1131. DOI:10.1111/1755-0998.12903 |
Liu J., Möller M., Provan J., et al, 2013. Geological and ecological factors drive cryptic speciation of yews in a biodiversity hotspot. New Phytol, 199: 1093-1108. DOI:10.1111/nph.12336 |
Liu Z.-F., Ci X.-Q., Li L., et al, 2017. DNA barcoding evaluation and implications for phylogenetic relationships in Lauraceae from China. PloS One, 12: e0175788. DOI:10.1371/journal.pone.0175788 |
Meeboonya R., Ngernsaengsaruay C., Balslev H., et al, 2019. The genus Dumasia(Fabaceae) in Thailand. Thai Forest Bull. Bot, 47: 113-120. DOI:10.20531/tfb.2019.47.1.15 |
Meier R., Shiyang K., Vaidya G., et al, 2006. DNA barcoding and taxonomy of Diptera: a tale of high intraspecific variability and low identification success. Syst. Biol, 55: 715-728. DOI:10.1080/10635150600969864 |
Merrill E.D., 1910. An enumeration of Philippine Leguminosae, with keys to the genera and species (concluded). Philipp. J. Sci, 5: 95-136. |
Meyer C.P., Paulay G., 2005. DNA barcoding: error rates based on comprehensive sampling. PLoS Biol, 3: e422. DOI:10.1371/journal.pbio.0030422 |
Möller M., Gao L.M., Mill R.R., et al, 2013. A multidisciplinary approach reveals hidden taxonomic diversity in the morphologically challenging Taxus wallichiana complex. Taxon, 62: 1161-1177. DOI:10.12705/626.9 |
Moore M.J., Soltis P.S., Bell C.D., et al, 2010. Phylogenetic analysis of 83 plastid genes further resolves the early diversification of eudicots. Proc. Natl. Acad. Sci.U.S.A, 107: 4623-4628. DOI:10.1073/pnas.0907801107 |
Newmaster S.G., Ragupathy S., 2009. Testing plant barcoding in a sister species complex of pantropical Acacia (Mimosoideae, Fabaceae). Mol. Ecol. Res, 9: 172-180. DOI:10.1111/j.1755-0998.2009.02642.x |
Pfenninger M., Nowak C., Kley C., et al, 2007. Utility of DNA taxonomy and barcoding for the inference of larval community structure in morphologically cryptic Chironomus (Diptera) species. Mol. Ecol, 16: 1957-1968. DOI:10.1111/j.1365-294X.2006.03136.x |
Posada D., Crandell K.A., 1998. Modeltest: testing the model of DNA substitution. Bioinformation, 14: 817-818. DOI:10.1093/bioinformatics/14.9.817 |
Posada D., Buckley T.R., 2004. Model selection and model averaging in phylogenetics: advantages of Akaike information criterion and Bayesian approaches over likelihood ratio tests. Syst. Biol, 53: 793-808. DOI:10.1080/10635150490522304 |
Pan B., Zhu X.-Y., 2010. Taxonomic revision of Dumasia (Fabaceae, Papilionoideae). Ann. Bot. Fenn, 47: 241-256. DOI:10.5735/085.047.0401 |
Pradeep S.V., Nayar M.P., 1991. Novelties in the genus Dumasia DC. (Leguminosae-Papilionoideae). J. Jpn. Bot, 66: 275-279. |
Ronquist F., Teslenko M., Van der Mark P., et al, 2012. MrBayes 3. 2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol, 61: 539-542. DOI:10.1093/sysbio/sys029 |
Sa, R., Gilbert, M.G., 2010. Dumasia DC. In: Wu, Z.-Y., Raven, P.H., Hong, D.-Y. (Eds.), Flora of China, vol. 10. Science Press, Beijing and Missouri Botanical Garden Press, St Louis, pp. 242-244.
|
Soltis D.E., Soltis P.S., Chase M.W., et al, 2000. Angiosperm phylogeny inferred from 18S rDNA, rbcL, and atpB sequences. Bot. J. Linn. Soc, 133: 381-461. DOI:10.1006/bojl.2000.0380 |
Stamatakis A., 2006. RAxML-VI-HPC: maximum likelihood-based phylogenetic analysis with thousands of taxa and mixed models. Bioinformatics, 22: 2688-2690. DOI:10.1093/bioinformatics/btl446 |
Struck T.H., Feder J.L., Bendiksby M., et al, 2018. Finding evolutionary processes hidden in cryptic species. Trends Ecol. Evol, 33: 153-163. DOI:10.1016/j.tree.2017.11.007 |
Taberlet P., Coissac E., Pompanon F., et al, 2007. Power and limitations of the chloroplast trnL (UAA) intron for plant DNA barcoding. Nucleic Acids Res, 35: e14. DOI:10.1093/nar/gkl938 |
Tyagi K., Kumar V., Singha D., et al, 2017. DNA Barcoding studies on thrips in India:cryptic species and Species complexes. Sci. Rep, 7: 4898. DOI:10.1038/s41598-017-05112-7 |
Tyagi K., Kumar V., Kundu S., et al, 2019. Identification of Indian Spiders through DNA barcoding: cryptic species and species complex. Sci. Rep, 9: 14033. DOI:10.1038/s41598-019-50510-8 |
Valentini A., Pompanon F., Taberlet P., 2009. DNA barcoding for ecologists. Trends Ecol. Evol, 24: 110-117. DOI:10.1016/j.tree.2008.09.011 |
Wei Y.-T., Lee S.-K., 1985. New material for Chinese leguminosae. Guihaia, 5: 157-174. |
Witt J.D.S., Threloff D.L., Hebert P.D.N., 2006. DNA barcoding reveals extraordinary cryptic diversity in an amphipod genus: implications for desert spring conservation. Mol. Ecol, 15: 3073-3082. DOI:10.1111/j.1365-294X.2006.02999.x |
Zhang J., Chen M., Dong X., et al, 2015. Evaluation of four commonly used DNA barcoding loci for Chinese medicinal plants of the family Schisandraceae. PloS One, 10: e0125574. DOI:10.1371/journal.pone.0125574 |



