b. University of Chinese Academy of Sciences, Beijing, 100049, China;
c. Center of Economic Botany, Core Botanical Gardens, Chinese Academy of Sciences, Menglun, Mengla, 666303, Yunnan, China;
d. Yunnan Key Laboratory for Rice Genetic Improvement, Food Crops Research Institute, Yunnan Academy of Agricultural Sciences, Kunming, 650200, Yunnan, China
DJY1 Dianjingyou 1
IL Introgression line
QTL Quantitative trait loci
LOD Logarithm of odds
GL Grain length
GNPP Grain number per panicle
GW Grain width
PB Primary branch number
PF Pollen fertility
PH Plant height
PL Panicle length
SB Secondary branch number
SF Spikelet fertility
SLL Sterile lemma length
SSR Simple sequence repeat
TGW 1000-grain weight
1. IntroductionRice (Oryza sativa L.) is a leading staple worldwide. Global food demands require the cultivation of high-quality, high-yield rice varieties that are resistant to multiple diseases. However, rice domestication resulted in a loss of genetic diversity, and rice yields have stagnated since the 1980s (Tanksley and Mccouch, 1997). Two major breakthroughs in rice yields have been facilitated by dwarf breeding and the generation of interspecific hybrids (Cheng et al., 2007; Peng et al., 2008). To increase genetic diversity of rice, breeders have focused on identifying genes responsible for desirable traits in relatives of cultivated rice varieties and transferring those genes to O. sativa (Xiao et al., 1998). However, reproductive barriers, such as hybrid sterility (Ouyang and Zhang, 2013), and linkage drag (Olsen et al., 2006; Palaisa et al., 2004) have prevented the transfer of desirable traits between cultivars and wild relatives.
Oryza glaberrima Steud. shares the same AA genome as O. sativa despite being derived from different ancestors (Pental and Barnes, 1985). O. glaberrima has numerous favorable agronomic traits, including resistance to biotic and abiotic stressors (Attere, 1983; Ghesquière et al., 1997). Moreover, O. glaberrima is considered an excellent gene pool for improving O. sativa (Xu et al., 2005). Hence, O. glaberrima yield-related genes may be used to break yield bottlenecks of O. sativa. One limitation to this approach is the hybrid incompatibility between O. sativa and O. glaberrima, which is attributed to postzygotic reproductive isolation leading to hybrid sterility (i.e., low fertility of hybrid progeny) (Ouyang and Zhang, 2013).
To date, more than 50 hybrid sterility loci have been identified in Oryza, including about 30 major and minor sterility loci (Ouyang and Zhang, 2013). Among these, 11 genes (Sa, S5, Sc, DPL1/DPL2, hsa1, S27/S28, S1, S7, DGS1/DGS2, qHMS7, ESA1) have been cloned that regulate the sterility of interspecific and indica-japonica intersubspecific hybrid sterility loci (Xie et al., 2019). Elucidating the regulatory mechanisms that control reproductive isolation between these rice species is critical for overcoming the incompatibility of interspecific hybridization that hinders the transfer of favorable genes across species.
Genes introduced into cultivated plants by backcross breeding programs are flanked by introgression segments of chromosomes derived from the donor parents. This process is commonly accompanied by linkage drag, in which traits other than those originally targeted are affected (Young and Tanksley, 1989). Linkage drag is intricately related to population structure, artificial selection, and genetic drift; furthermore, the intensity of linkage drag is tightly dependent on the physical distance of linked genes (Liu et al., 2009). Linkage intensity has been estimated and used to study genetic patterns of linkage drag in barley (Brown et al., 1989). In rice, blast-resistance is controlled by Pi-zt, which is located on the short arm of chromosome 6 and is tightly linked with late maturity in the progenies of different indica and japonica crosses (Yokoo and Fujimaki, 1971). The negative association between the rice blast-resistance gene pi25(t) and a yield-related quantitative trait loci (QTLs) made it difficult to breed both high resistance and high yield progenies (Zhuang et al., 2001). In addition, some traits show linkage drag without recombination in the process of long-term artificial selection and domestication. For instance, selection for alleles associated with larger fruits alter metabolite profiles as a consequence of linkage with nearby genes in tomato (Zhu et al., 2018). Long-range patterns of diversity and linkage disequilibrium surrounding the maize Y1 gene indicate an asymmetric selective sweep (Palaisa et al., 2004). Moreover, even in cases where backcross breeding has been repeated many times, donor parent chromosome segments up to 10 cM have been found around the target gene, although genetic recombination rarely occurred (Tanksley, 1993). To date, linkage drag has been well documented in crop breeding and domestication analysis that cannot be explained by linkage intensity. Thus, the underlying causes of linkage drag have yet to be elucidated.
In this study, we dissected an introgression segment in a high generation backcross introgression line (BC4F10) by using O. glaberrima as the donor in a japonica rice background. We mapped a hybrid sterility locus, S20, a long sterile lemma locus, G1-g, and a grain width QTL, qGW7, to a 15 cM-linkage region. S20 eliminated the male gametes of O. sativa, which led all hybrid progenies to have homozygous of O. glaberrima genotypes. The proportion of the differentiation genotypes of the fertility genes deviated from Mendelian segregation ratios. The same segregation distortion was observed for the linked traits G1-g and qGW7, implying an association with S20 that leads to linkage drag.
2. Materials and methods 2.1. Materials and growthTo raise a set of BC4 introgression lines (ILs), we used Dianjingyou 1 (DJY1), an elite japonica cultivar from Yunnan province (China), as the recurrent parent, and IRGC102555, an accession of O. glaberrima introduced from the International Rice Research Institute (IRRI), as the donor parent. To obtain BC5F1, IL-2769 (BC4F10) was backcrossed with DJY1 (Chen et al., 2018). The BC5F2 population was obtained from BC5F1 plants at the breeding base of Xishuangbanna Tropical Botanical Garden (XTBG), Mengla county, Yunnan province, China. DJY1, IL-2769, BC5F1, and BC5F2 populations were sown on seedbeds. When grown to quatrefoil stage, seedlings were transplanted to a field with 25 × 20 cm between plants. Field management was carried out in accordance with general rice production.
2.2. Morphological analysisWe measured the following agronomic traits of DJY1, IL-2769, BC5F1, and BC5F2 populations: plant height, panicle length, sterile lemma length, grain length, grain width, primary and secondary branch number, grain number per panicle, 1000-grain weight, pollen and spikelet fertility.
The BC5F1 and BC5F2 individuals were each used for genetic analysis. Pollen fertility was determined as described by Doi et al. (1998). Briefly, the panicles were sampled before anthesis and fixed in 70% ethanol, and then stained by I2-KI 1% solution. Pollen grains were classified as fertile or sterile. Sterile pollen was further classified as unstained withered sterile, unstained spherical sterile, and stained abortive (Li, 1980). At least three independent microscopic fields were viewed to calculate the ratio of fertile pollen grains.
2.3. DNA extraction and SSR analysisDNA extraction was based on the cetyltrimethylammonium bromide (CTAB) method (Ling et al., 2018). A total of 450 simple sequence repeats (SSR), covering most of the rice genome, were used to screen polymorphisms between DJY1 and IL-2769. PCR amplifications were performed in a total reaction volume of 10 μL containing 10 ng DNA template, 1 × buffer, 0.5 μmol/L of each primer, 50 μmol/L of dNTPs and 0.5 U of Taq polymerase. PCR amplification was performed under the following conditions: pre-denaturation at 94 ℃ for 3 min, 29 cycles of denaturation at 94 ℃ for 30 s, renaturation at 62 ℃ and extension at 72 ℃ for 25 s, followed by a final extension at 72 ℃ for 5 min. PCR products were visualized on 10% non-denatured polyacrylamide gel stained with silver nitrate (Bassam et al., 1991).
2.4. Linkage mapLinkage maps of qualitative traits and markers were developed using the MAPMAKER EXP 3.0 (Lander et al., 1987). The QTL IciMapping V4.2 at logarithm of odds (LOD) scores > 2.5 was used for quantitative trait detection (Li et al., 2007). The Kosambi function was used to transform recombination frequencies of genetic distances. The genetic map was drawn using the Mapchart 2.32 (Voorrips, 2002).
2.5. Statistical analysisThe results are means ± standard deviation (SD). Statistical analysis was performed by IBM SPSS Statistics 20. Student's t-tests were used for analysis the difference between the progenies and their parents.
3. Results 3.1. Phenotype comparison between the introgression line IL-2769 and the receptor parent DJY1Phenotype analysis of the IL-2769 and DJY1 showed significant or highly significant differences in several traits, including panicle length, secondary branch number, sterile lemma length, grain length, grain width, 1000-grain weight, grain number per panicle, plant height, spikelet fertility, and primary branch number, but not in pollen fertility (Table 1). Although the theoretical introgression ratio of the BC4 introgression line was 3.125%, there were still many significant differences between IL-2769 and the recurrent parent, implying that linkage drag might contribute to these differences.
| Lines | BC5F1 | IL-2769 | DJY1 |
| Pollen fertility (%) | 49.80 ± 0.63** | 94.90 ± 2.93 | 96.70 ± 1.59 |
| Spikelet fertility (%) | 88.40 ± 3.63 | 89.50 ± 3.73 | 88.00 ± 2.38 |
| Panicle length (cm) | 22.14 ± 1.74 | 23.10 ± 1.73** | 21.32 ± 1.32 |
| Primary branch number | 10.50 ± 2.72* | 7.50 ± 1.04 | 8.40 ± 0.82 |
| Secondary branch number | 14.60 ± 7.43 | 7.90 ± 3.71** | 14.50 ± 4.14 |
| Sterile lemma length (mm) | 4.65 ± 0.41** | 7.52 ± 0.19** | 2.83 ± 0.20 |
| Grain length (mm) | 9.32 ± 0.74 | 8.83 ± 0.19* | 9.23 ± 0.22 |
| Grain width (mm) | 3.90 ± 0.33 | 4.29 ± 0.10** | 3.76 ± 0.07 |
| 1000-grain weight (g) | 30.48 ± 1.62* | 30.21 ± 0.50* | 28.19 ± 0.47 |
| Grain number per panicle | 73.20 ± 6.03** | 68.30 ± 14.87** | 91.30 ± 15.25 |
| Plant height (cm) | 98.4 ± 2.46* | 101.81 ± 3.87** | 94.57 ± 3.52 |
| * and ** indicate significant differences from control at p < 0.05 and p < 0.01, respectively; the BC5F1 and IL-2769 columns are marked in comparison with DJY1. | |||
To analyze the genetic basis of traits carried by the IL-2769, we crossed IL-2769 plants with DJY1. The hybrid (BC5F1) pollens were semi-sterile, showing stained abortive grains; however, spikelets were fertile. The BC5F1 hybrids showed phenotypes that were intermediate between or similar to their parents for the following traits: sterile lemma length, grain width, panicle length, primary branch number, secondary branch number, 1000-grain weight, grain number per panicle and plant height (Table 1, Fig. 1). These findings indicate that these traits are semi dominant in DJY1 or controlled by quantitative loci.
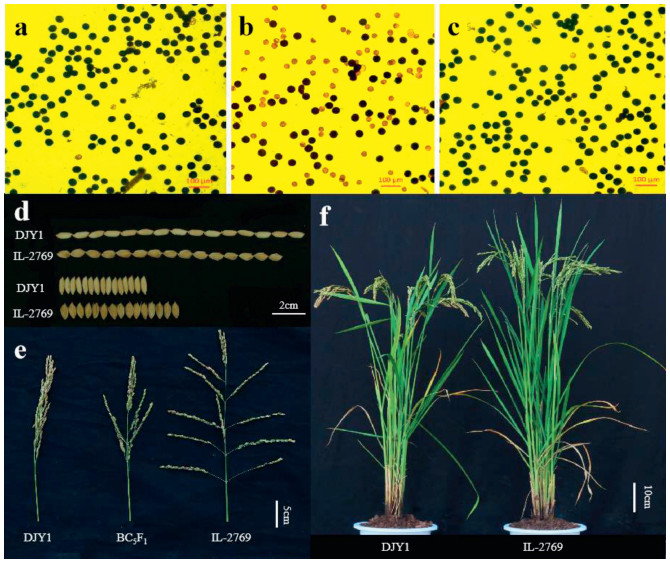
|
| Fig. 1 Phenotypes of BC5F1, DJY1 and IL-2769. (a–c) Pollen fertility of DJY1, BC5F1 hybrid and IL-2769, respectively; (d) Grain comparison of DJY1 and IL-2769 (n = 15); (e) Panicle type of DJY1, BC5F1 hybrid and IL-2769; (f) Plant architecture of two parents. |
In the BC5F2 population, pollen fertility and sterile lemma length showed a clear-cut bimodal distribution. The phenotypic ratio of hybrid individuals with semi-sterile pollen (about 50% fertility) and those with fertile pollen (about 100% fertility) was 97:81, which is consistent with a 1:1 ratio rather than of the normal 3:1 segregation ratio (χ2 = 0.721, P = 0.296; Fig. 2). Taken together, pollen semi-sterility and segregation distortion in BC5F1 suggest that the male gametes may have been eliminated. The long and short sterile lemma groups also produced a 1:1 ratio (90:88, χ2 = 0.011, P = 0.916), which implies that sterile lemmas were simultaneously affected by male gamete elimination. These results suggest that pollen grain fertility and sterile lemma are controlled by one locus. Grain width showed continuous bimodal distribution. Panicle length, primary branch number, secondary branch number, grain length and 1000-grain weight presented a similar continuous distribution (Fig. 2).
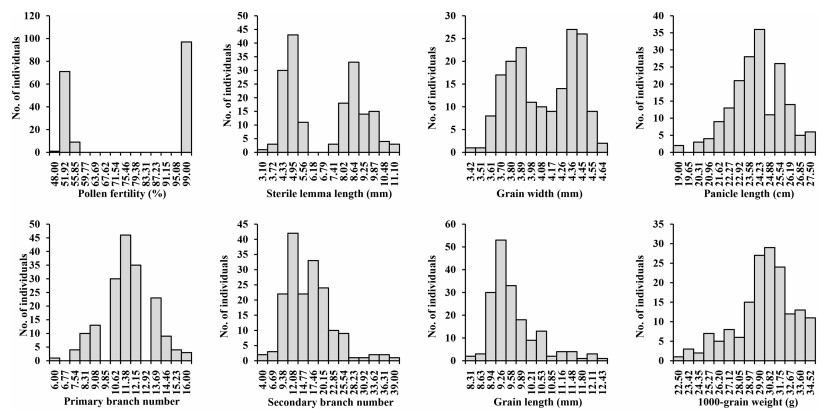
|
| Fig. 2 Distribution of traits in BC5F2 population. |
Correlation analysis of traits showed that pollen fertility, sterile lemma length, grain width, grain length and 1000-grain weight were significantly correlated with each other; and panicle length, primary branch number, secondary branch number were also significantly correlated with each other. The primary branch number and secondary branch number were significantly correlated with grain width, grain length, and 1000-grain weight (Table 2). These results suggest that the related traits might be clustered on the same chromosomes.
| Traits | PF | SF | PL | PBN | SBN | SLL | GL | GW | TGW |
| PF | 1 | ||||||||
| SF | 0.789** | 1 | |||||||
| PL | 0.070 | 0.069 | 1 | ||||||
| PBN | -0.072 | 0.014 | 0.645** | 1 | |||||
| SBN | -0.114 | -0.007 | 0.437** | 0.567** | 1 | ||||
| SLL | 0.811** | 0.702** | 0.045 | -0.108 | -0.067 | 1 | |||
| GL | 0.207** | 0.017 | 0.126 | -0.180* | -0.278** | 0.331** | 1 | ||
| GW | 0.750** | 0.679** | 0.030 | -0.169* | -0.096 | 0.902** | 0.227** | 1 | |
| TGW | 0.187* | 0.133 | 0.064 | -0.178* | -0.183* | 0.204** | 0.460** | 0.321** | 1 |
| PF: Pollen fertility (%); SF: Spikelet fertility (%); PL: Panicle length (cm); PB: Primary branch number; SB: Secondary branch number; SLL: Sterile lemma length (mm); GL: Grain length (mm); GW: Grain width (mm); TGW: 1000-grain weight (g); GNPP: Grain number per panicle; PH: Plant height (cm); * and ** indicate significant correlations at p < 0.05 and p < 0.01, respectively. | |||||||||
A total of 450 SSR markers with polymorphisms between the two parents, DJY1 and IRGC102555, were used to survey the introgression segments in IL-2796. We found a 15-cM introgression segment on the short arm of chromosome 7 and used 13 SSR markers to screen the introgression region for genotypes of 178 BC5F2 individuals. Based on the phenotypes and genotypes of BC5F2 individuals, two loci and a QTL from O. glaberrima were mapped to the introgression region (Fig. 3). A hybrid sterility locus was restricted to a 1.7-cM region flanked by RM20866 and RM20896, and co-segregated with SSR marker RM20894. This mapping region was similar to the region of S20 identified in a cross between O. sativa and O. glaberrima (Doi et al., 1999); hence it was named S20. The long sterile lemma locus was located at a 1.2-cM region flanked by RM20943 and RM5752, which was adjacent to long sterile lemma (G1) (Yoshida et al., 2009; Liu et al., 2016), and was designated as G1-g as it came from O. glaberrima. In addition, we mapped a new grain width QTL (qGW7) flanked by RM20866 and RM20933, which may explain 39.3% of phenotype variation (Table 3).
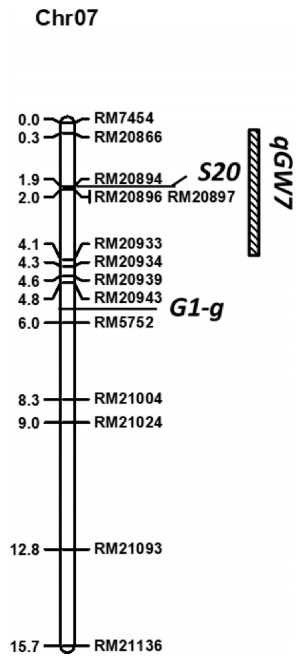
|
| Fig. 3 The positions of linkage traits in the introgression region at the end of the short arm of chromosome 7. |
| QTL | Marker Interval | LOD | A | D | R2 (%) |
| qGW7 | RM20866-RM20933 | 56.58 | 0.32 | 0.18 | 39.33 |
| A: Additive effect, D: Dominant effect, allele from O. glaberrima; R2: Proportion of the phenotypic variance explained by the QTL. | |||||
We further analyzed genotype segregation ratios by SSR markers closely linked to mapping traits. RM20894 co-segregated with S20 and their offspring's genotypes were homozygotes of O. glaberrima (GG) and heterozygotes (GS), but not homozygotes of O. sativa (SS). The pollen grains of GG homozygotes showed normal pollen fertility, and heterozygotes exhibited semi-sterility. These results indicate that the interaction between S20-g and S20-s led to the complete abortion of the male gametes carrying the allele of S20-s in the heterozygotes, which resulted in segregation distortion in BC5F2 population. Similarly, G1-g and qGW7, which were tightly linked to marker RM5752 and RM20866, respectively, also showed segregation distortion (Table 4). Likewise, the O. sativa homozygous genotypes of linked G1-g and qGW7 were seldom produced in BC5F2 progenies, and the homozygous GG and heterozygous produced an approximately 1:1 segregation ratio in BC5F2 progenies. The homozygous GG genotypes showed genetic stability, and potentially reduced the frequency of chromosome exchange in progenies. These results indicate that S20 led to linkage drag of traits in the introgression segment from O. glaberrima.
| Loci | Linkage markers | No. of genotype | χ2 (1:2:1) | P | ||
| SS | SG | GG | ||||
| S20 | RM20894 | 0 | 97 | 81 | 75.16 | 4.78409E-17 |
| G1-g | RM5752 | 5 | 86 | 87 | 75.75 | 3.55211E-17 |
| GW7 | RM20866 | 1 | 95 | 82 | 74.53 | 6.55286E-17 |
| SS, SG and GG indicate DJY1-homozygous, heterozygous and O. glaberrima-homozygous genotypes, respectively. | ||||||
More than 50 hybrid sterility loci from Oryza have been identified. These interspecific and indica-japonica intersubspecific hybrid sterility loci are distributed on most rice chromosomes (Ouyang et al., 2009). Most of them follow the one-locus allelic interaction model (Kitamura, 1962). The genetic basis for hybrid sterility in rice has been divided into two types: pollen killers and gamete eliminators (Sano, 1990). A single pollen killer locus produces segregation ratios of 1:1, consisting of heterozygotes and homozygotes of only one parent; in contrast, a gamete eliminator locus produces only individuals homozygous for one parent (Rick, 1966). In this study, we detected linkage drag caused by a hybrid sterility locus, which we call S20 because it is located in a region similar to that of the S20 previously identified in a cross between O. sativa and O. glaberrima (Doi et al., 1999). Widely distributed hybrid sterility loci have the same effect on the linkage traits as S20. Therefore, we speculate that these loci are the critical factors for the formation of linkage drag.
Transferring favorable alleles into O. sativa from its wild relatives will be beneficial for crop breeding. Hybrid sterility is the main reproductive barrier. Most hybrid sterility loci that have been detected are derived from wild relatives and can eliminate the gametes of O. sativa. Linkage drag may be caused by sterility loci that hinder the production of recombinant progenies with desirable traits and increase obstacles during breeding. Some loci associated with yield, resistance and other favorable traits are linked with hybrid sterility loci. A number of hybrid sterility loci have been extensively studied, including S10 with Grain Size 6 (GS6) (Sun et al., 2013), S27 with PLANT ARCHITECTURE AND YIELD 1 (PAY1) (Zhao et al., 2015), S31 with small and round seed 3 (SRS3) and grain size 5 (GS5) (Kitagawa et al., 2010; Li et al., 2011). In our study, we also observed segregation distortion (likely influenced by S20) for G1-g, which is located within a 1.2-cM region flanked by RM20943 and RM5752, and is adjacent to long sterile lemma (G1) (Yoshida et al., 2009; Liu et al., 2016). Another linked trait, qGW7, produces similar segregation distortion. These results imply that linkage drag occurred during interspecific hybridization.
Hence, it is increasingly urgent to find strategies that break interspecific hybrid sterility and linkage drag. Recent studies have shown that exploring or constructing neutral wide-compatibility loci in natural variation, or using near isogenic lines of hybrid sterility genes could bridge the gaps between parents and offspring and further break hybrid sterility (Koide et al., 2018; Li et al., 2018), which could provide new insights into breaking the linkage drag efficiently.
Author contributionsConceptualization, P.X. and D.Y.; methodology, D.T. and P.X.; validation, M.W. and J.Z.; formal analysis, J.Z. and J.W.; investigation, M.W. and J.Y.; resources, D.T. and P.X.; data curation, M.W.; writing-original draft preparation, M.W.; writing-review and editing, P.X. and J.W.; supervision, P.X. and D.Y.; funding acquisition, P.X. and D.Y. All authors have read and agreed to the published version of the manuscript.
Declaration of Competing InterestThe authors declare that they have no conflict of interest.
AcknowledgmentsThe authors thank the Public Technology Service Center, Xishuangbanna Tropical Botanical Garden, Chinese Academy of Sciences for technical support. This work was supported by "One-Three-Five" Strategic Planning of Chinese Academy of Sciences (2017XTBG-T02) and Strategic Leading Science and Technology Program (XDA24030301 and XDA24040308).
Attere A.F., 1983. Reaction of Oryza glaberrima accessions to rice yellow mottle virus. Plant Dis, 67: 420-421. DOI:10.1094/PD-67-420 |
Bassam B.J., Caetano-Anollés G., Gresshoff P.M., 1991. Fast and sensitive silver staining of DNA in polyacrylamide gels. Anal. Biochem, 196: 80-83. DOI:10.1016/0003-2697(91)90120-I |
Brown A.H.D., Lawrence G.J., Jenkin M., et al, 1989. Linkage drag in backcross breeding in barley. J. Hered, 80: 234-239. DOI:10.1093/oxfordjournals.jhered.a110841 |
Chen X.Q., Xu P., Zhou J.W., et al, 2018. Mapping and breeding value evaluation of a semi-dominant semi-dwarf gene in upland rice. Plant Divers, 40: 238-244. DOI:10.1016/j.pld.2018.09.001 |
Cheng S.H., Zhuang J.Y., Fan Y.Y., et al, 2007. Progress in research and development on hybrid rice: a super-domesticate in China. Ann. Bot, 100: 959-966. DOI:10.1093/aob/mcm121 |
Doi K., Yoshimura A., Iwata N., 1998. RFLP mapping and QTL analysis of heading date and pollen sterility using backcross populations between Oryza sativa L. and Oryza glaberrima. Steud. Breed Sci, 48: 395-399. |
Doi K., Taguchi K., Yoshimura A., 1999. RFLP mapping of S20 and S21 for F1 pollen semi-sterility found in backcross progeny of Oryza sativa and O. glaberrima. Rice Genet. Newsl, 16: 65-68. |
Ghesquière A., Séquier J., Second G., et al, 1997. First steps towards a rational use of African rice, Oryza glaberrima, in rice breeding through a 'contig line' concept. Euphytica, 96: 31-39. DOI:10.1023/A:1003045518236 |
Kitagawa K., Kurinami S., Oki K., et al, 2010. A novel kinesin 13 protein regulating rice seed length. Plant Cell Physiol, 51: 1315-1329. DOI:10.1093/pcp/pcq092 |
Kitamura E., 1962. Studies on cytoplasmic sterility of hybrids in distantly related varieties of rice, (Oryza sativa L.). Jap. J. Breed, 2: 81-84. |
Koide Y., Ogino A., Yoshikawa T., et al, 2018. Lineage-specific gene acquisition or loss is involved in interspecific hybrid sterility in rice. Proc. Natl. Acad. Sci. U.S.A, 115: E1955-E1962. |
Lander E.S., Green P., Abrahamson J., et al, 1987. MAPMAKER: an interactive computer package for constructing primary genetic linkage maps of experimental and natural populations. Genomics, 93: 398. |
Li H., Ye G., Wang J., 2007. A modified algorithm for the improvement of composite interval mapping. Genetics, 175: 361-374. DOI:10.1534/genetics.106.066811 |
Li J., Zhou J.W., Xu P., et al, 2018. Neutral alleles at hybrid sterility loci of Oryza glaberrima from AA genome relatives in genus Oryza. Breed Sci, 68: 343-351. DOI:10.1270/jsbbs.18006 |
Li Y.B., Fan C.C., Xing Y.Z., et al, 2011. Natural variation in GS5 plays an important role in regulating grain size and yield in rice. Nat. Genet, 43: 1266-1269. DOI:10.1038/ng.977 |
Li Z.B., 1980. A preliminary discussion about the classification of male sterile lines of rice in China. Acta Agron. Sin, 6: 17-26. |
Ling F., Wu H., Lin Y.J., et al, 2018. A rapid method for rice DNA extraction. Bio-101: e1010101. |
Liu M.J., Li H.F., Su Y.L., et al, 2016. G1/ELE functions in the development of rice lemmas in addition to determining identities of empty glumes. Front. Plant Sci, 7: 1006. |
Liu W.Q., Fan Y.Y., Chen J., et al, 2009. Avoidance of linkage drag between blast resistance gene and the QTL conditioning spikelet fertility based on genotype selection against heading date in rice. Rice Sci, 16: 21-26. DOI:10.1016/S1672-6308(08)60052-9 |
Olsen K.M., Caicedo A.L., Polato N., et al, 2006. Selection under domestication:evidence for a sweep in the rice waxy genomic region. Genetics, 173: 975-983. DOI:10.1534/genetics.106.056473 |
Ouyang Y.D., Chen J.J., Ding J.H., et al, 2009. Advances in the understanding of inter-subspecific hybrid sterility and wide-compatibility in rice. Chin. Sci. Bull, 54: 2332-2341. DOI:10.1007/s11434-009-0371-4 |
Ouyang Y.D., Zhang Q.F., 2013. Understanding reproductive isolation based on the rice model. Annu. Rev. Plant Biol, 64: 111-135. DOI:10.1146/annurev-arplant-050312-120205 |
Palaisa K., Morgante M., Tingey S., et al, 2004. Long-range patterns of diversity and linkage disequilibrium surrounding the maize Y1 gene are indicative of an asymmetric selective sweep. Proc. Natl. Acad. Sci. U.S.A, 101: 9885-9890. DOI:10.1073/pnas.0307839101 |
Peng S., Khush G.S., Virk P., et al, 2008. Progress in ideotype breeding to increase rice yield potential. Field Crop. Res, 108: 32-38. DOI:10.1016/j.fcr.2008.04.001 |
Pental D., Barnes S.R., 1985. Interrelationship of cultivated rices Oryza sativa and O. glaberrima with wild O. perennis complex. Theor. Appl. Genet, 70: 185-191. DOI:10.1007/BF00275320 |
Rick C.M., 1966. Abortion of male and female gametes in the tomato determined by allelic interaction. Genetics, 53: 85-96. |
Sano Y., 1990. The genic nature of gamete eliminator in rice. Genetics, 125: 183-191. |
Sun L.J., Li X.J., Fu Y.C., et al, 2013. GS6, a member of the GRAS gene family, negatively regulates grain size in rice. J. Integr. Plant Biol, 55: 938-949. |
Tanksley S.D., 1993. Mapping polygenes. Annu. Rev. Genet, 27: 205-233. DOI:10.1146/annurev.ge.27.120193.001225 |
Tanksley S.D., Mccouch S.R., 1997. Seed banks and molecular maps: unlocking genetic potential from the wild. Science, 277: 1063-1066. DOI:10.1126/science.277.5329.1063 |
Voorrips R.E., 2002. MapChart: software for the graphical presentation of linkage maps and QTLs. J. Hered, 93: 77-78. DOI:10.1093/jhered/93.1.77 |
Xiao J., Li J., Grandillo S., et al, 1998. Identification of trait-improving quantitative trait loci alleles from a wild rice relative. Oryza rufipogon. Genetics, 150: 899-909. |
Xie Y.Y., Shen R.X., Chen L.T., et al, 2019. Molecular mechanisms of hybrid sterility in rice. Sci. China Life Sci, 62: 737-743. DOI:10.1007/s11427-019-9531-7 |
Xu P., Tao D.Y., Hu F.Y., et al, 2005. Interspecific hybridization of cultivated rice for breeding japonica rice in Yunnan province. Chin. J. Rice Sci, 19: 41-46. |
Yokoo M., Fujimaki H., 1971. Tight linkage of blast-resistance with late maturity observed in different indica varieties of rice. Jpn. J. Breed, 21: 35-39. DOI:10.1270/jsbbs1951.21.35 |
Yoshida A., Suzaki T., Tanaka W., et al, 2009. The homeotic gene long sterile lemma(G1) specifies sterile lemma identity in the rice spikelet. Proc. Natl. Acad. Sci.U.S.A, 106: 20103-20108. DOI:10.1073/pnas.0907896106 |
Young N.D., Tanksley S.D., 1989. RFLP analysis of the size of chromosomal segments retained around the Tm-2 locus of tomato during backcross breeding. Theor.Appl. Genet, 77: 353-359. DOI:10.1007/BF00305828 |
Zhao L., Tan L.B., Zhu Z.F., et al, 2015. PAY1 improves plant architecture and enhances grain yield in rice. Plant J, 83: 528-536. DOI:10.1111/tpj.12905 |
Zhu G., Wang S., Huang Z., et al, 2018. Rewiring of the fruit metabolome in tomato breeding. Cell, 172: 249-261 e12. DOI:10.1016/j.cell.2017.12.019 |
Zhuang J.Y., Wu J.L., Fan Y.Y., et al, 2001. Genetic drag between a blast resistance gene and QTL conditioning yield trait detected in a recombinant inbred line population in rice. Rice Genet. Newsl, 18: 69-70. |



