b. Yunnan Key Laboratory for Wild Plant Resources, Kunming, Yunnan, China;
c. Key Laboratory of Tropical Forest Ecology, Xishuangbanna Tropical Botanical Garden, Chinese Academy of Sciences, Mengla, Yunnan, China;
d. Flower Research Institute, Yunnan Academy of Agricultural Sciences, Kunming, Yunnan, 650205, China
More than 70% of orchids are epiphytic or lithophytic, having evolved from their terrestrial ancestors to adapt to environments that differ in terms of solar radiation, as well as nutrient and water supply (Gravendeel et al., 2004). Orchids with different life forms are characterized by different adaptive structures and functional traits (Zhang et al., 2018), but all have mycorrhizal fungal associations that are very important for their growth, from germination to adulthood (Yukawa et al., 2009; Dearnaley et al., 2012). Identifying the community composition of orchid mycorrhizal fungi (OMF) is essential to understanding their biotic mutualisms with orchid species, and the factors influencing OMF composition. High-throughput sequencing produces a great deal of data and has significantly enhanced the efficiency of identifying OMF and comparing different OMF communities (Waud et al., 2014; Jacquemyn et al., 2017). A previous study has reported OMF have highly modular architecture, reflecting an ecological barrier between epiphytic and terrestrial subnetworks on a paleotropic island (Martos et al., 2012). Other studies have also reported OMF specificities (mostly Sebacinales and Tulasnellaceae), differing between terrestrial, epiphytic, and lithophytic habitats (Oja et al., 2015; Těšitelová et al., 2015; Xing et al., 2015, 2019). Because studies on the relationships among OMF compositions and orchid life forms were all conducted in tropical areas, it remains unclear if individual epiphytic and lithophytic orchids harbor different OMF at higher elevations.
Coelogyne Lindl. (Epidendroideae, Orchidaceae) contains ca. 200 species that are distributed throughout tropical and subtropical parts of Asia to Oceania, with 31 species occurring in China (Gravendeel et al., 2001; Chen et al., 2009). Species within this genus retain only one or two leaves and cannot produce new leaves during the growing season. Coelogyne orchids can be both epiphytic and lithophytic at the same site and are always colonized by mycorrhizal fungi. Some Coelogyne species have therefore been studied to assess correlations of OMF composition and life form (Xing et al., 2015, 2019). Many Coelogyne species are also of high ornamental value, and wild populations of this genus have been threatened by over-collection and habitat destruction. Understanding their OMF would therefore be useful to support seedling propagation and artificial cultivation of these sought-after flower resources.
Studies on the OMF of Coelogyne orchids have been mainly conducted on two tropical species—Coelogyne viscosa Rchb. f. and Coelogyne ovalis Lindl., and have been based on Sanger sequencing of randomly picked clones from amplicon libraries of the ITS-rDNA region (Xing et al., 2015, 2019). The OMF Operational Taxonomic Units (OTUs) have been mostly identified as fungi of Sebacinales and Tulasnellaceae, with only a few OTUs designated as Ceratobasidiaceae. Mycorrhizal fungal community composition is significantly different between epiphytic and lithophytic individuals of C. viscosa (Xing et al., 2015). The habitat distribution of C. viscosa is restricted between 1000 m and 2000 m above sea level. In contrast, another species, C. corymbosa Lindl., can be found at an elevation of 3500 m. This is almost the highest elevation at which epiphytic orchids are found. C. corymbosa is also the species with the northern-most distribution within the genus. Given that mycorrhizal fungal species compositions can be affected by the geographic distribution of plant hosts (Gai et al., 2012; Gorzelak et al., 2012; Looney et al., 2016; Duffy et al., 2019), we speculated that there are differences in the OMF composition between C. corymbosa and its tropical relatives, and between epiphytic and lithophytic individuals of the species.
In an investigation of the plants of western Yunnan Province in China, four populations of C. corymbosa were found growing in habitats within subtropical monsoon or subalpine regions. We sampled the species' mycorrhizal fragments to detect its OMF diversity using high-throughput sequencing and to compare the OMF communities from the different plant populations. Our objectives were twofold: (ⅰ) Do the sampled C. corymbosa orchids have the same OMF as their tropical relatives? (ⅱ) Do OMF communities differ between litho- and epiphytic orchids of a certain site, or differ between samples of each collecting site?
2. Materials and methods 2.1. SamplingMycorrhizae of wild plant individuals of C. corymbosa (Fig. 1) were collected from three sites in western Yunnan Province, southwestern China, in May 2017. Both epiphytic and lithophytic plants were sampled from Longling County (LL, elev. 2500 m, 24°32′N, 98°53′E), while only lithophytic plants were found and sampled from Baoshan (BS, elev. 2500 m, 25°11′N, 98°59′E) and Dali (DL, elev. 2600 m, 25°38′N, 100°08′E). We sampled 6 individuals each (at least 3 root fragments for each individual) from BS and DL, 6 from rock surface and 6 from tree stems in LL. In total 72 root fragments from 24 individuals were obtained, and then merged into 12 samples (by mixing every two individuals) for high-throughput sequencing.

|
| Fig. 1 Wild plants of Coelogyne corymbosa and transection features of their mycorrhizae. A. Plants growing in lithophytic habitat. B-D. Brownish fungal pelotons colonized in mycorrhizae are indicated by blue arrows. |
Fresh orchid roots were rinsed with tap water and gently brushed to remove soil particles adhering to the surface of the root. Mycorrhizal colonization was confirmed by checking for the existence of fungal pelotons under a light microscope (Fig. 1). These root fragments were then surface-sterilized with NaClO/ethanol and rinsed three times in sterile water. Afterwards, they were frozen in liquid nitrogen and stored at -80 ℃ prior to molecular analyses.
2.2. Molecular sequencingThe frozen roots were ground to powder and total DNA was extracted with a Plant DNA Rapid Extraction Kit (BioTeke Beijing, China) according to the manufacturer's instructions. Amplicon libraries of ITS2-rDNA sequences were created using two primer pairs, ITS3/ITS4OF and ITS86F/ITS4 (Gardes et al., 1991; Turenne et al., 2000; Taylor and McCormick, 2008), which were recommended by previous studies (Waud et al., 2014, 2016) because of their good performance in OMF community identification. Polymerase chain reaction (PCR) amplification, purification, quantification, library construction, and sequencing (Illumina MiSeq PE-250) were conducted by Personalbio Tec. Co., Ltd. (Shanghai, China).
2.3. Data analysesSequences obtained were assigned to the appropriate sample based on both barcode and primer sequences, allowing zero discrepancies, and were subsequently trimmed from the barcodes and primers using CUTADAPT 1.0 (Martin, 2011). Sequences were trimmed based on a minimum Phred score of 30 (base call accuracy of 99.9%) averaged over a 50 bp moving window; short reads, chimeras, and singletons were then excluded in Usearch11 (Edgar and Flyvbjerg, 2015). After that, the ITS2 subregion of the sequences was extracted with ITSx v1.1 (Bengtsson-Palme et al., 2013) before being clustered into OTUs at 97% identity threshold using UPARSE-OTU algorithm (Edgar, 2013). The Blast (20171201) and UNITE databases were used for taxonomic assignments of the OTUs (Kõljalg et al., 2005).
The OTUs of Ceratobasidiaceae, Sebacinales, Tulasnellaceae, and Thelephoraceae were designated as putative OMF based on previously published definitions of orchid mycorrhizae (Dearnaley et al., 2012). Representative sequences for the putative OMF OTUs were deposited in GenBank (MT548941 through MT549013, see Supplementary Table 1). The OTUs of Cantharellales (including Ceratobasidiaceae and Tulasnellaceae) and Sebacinales (Serendipitaceae) were the most abundant. To understand relationships among the mycobionts detected from C. corymbosa and other congeneric species, phylogenetic analyses of the above mentioned two fungal lineages were conducted. Sequences were aligned using Mafft (Katoh and Standley, 2013) and manually checked with BioEdit 7.0.9 (Hall, 1999). Maximum Likelihood (ML) algorithm was then employed by using RAxML 7.2.6 (Stamatakis, 2006). GTR + I + G was selected as the best substitution model suite for the two data sets when applying the Akaike Information Criterion implemented in Mrmodeltest 2.3 (Nylander, 2004). All parameters in the ML analysis were kept at their default levels, and statistical support was obtained using a rapid nonparametric bootstrapping with 1000 replicates.
| Data source*distance | Factors | Mean squares | Variation (R2) | Pr (> F) |
| abundance*bray | site | 0.55 | 0.28 | 0.11 |
| life form | 0.20 | 0.05 | 0.80 | |
| presence*jaccard | site | 0.43 | 0.47 | 0.001 |
| life form | 0.18 | 0.10 | 0.34 |
Operational Taxonomic Unit (OTU) abundance data were generated from all the merged sequences and OTUs. This was then homogenized using rarefaction.py in Qiime 2 (Caporaso et al., 2010) according to the lowest sequence abundance (19493) of all OTUs for each sample. The homogenized OTU abundance data and the relevant binary data were used for Non-metric multidimensional scaling (NMDS) based on Bray-Curtis and Jaccard dissimilarity matrices in R package vegan (Oksanen et al., 2013). To test whether fungal composition differs between collection sites and life forms (i.e., epiphytic or lithophytic), a distance-based multivariate analysis of variance (Permanova) (Anderson et al., 2008) was also performed using the Adonis function in vegan. Afterwards, Venn diagrams were used to visualize the OMF composition of each plant population.
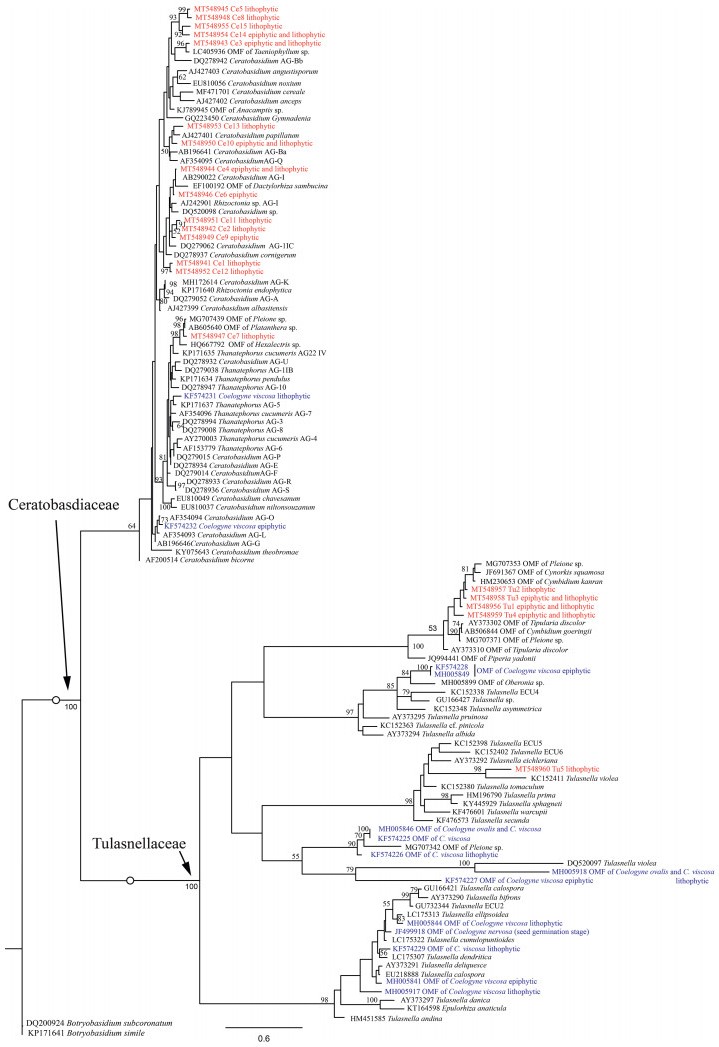
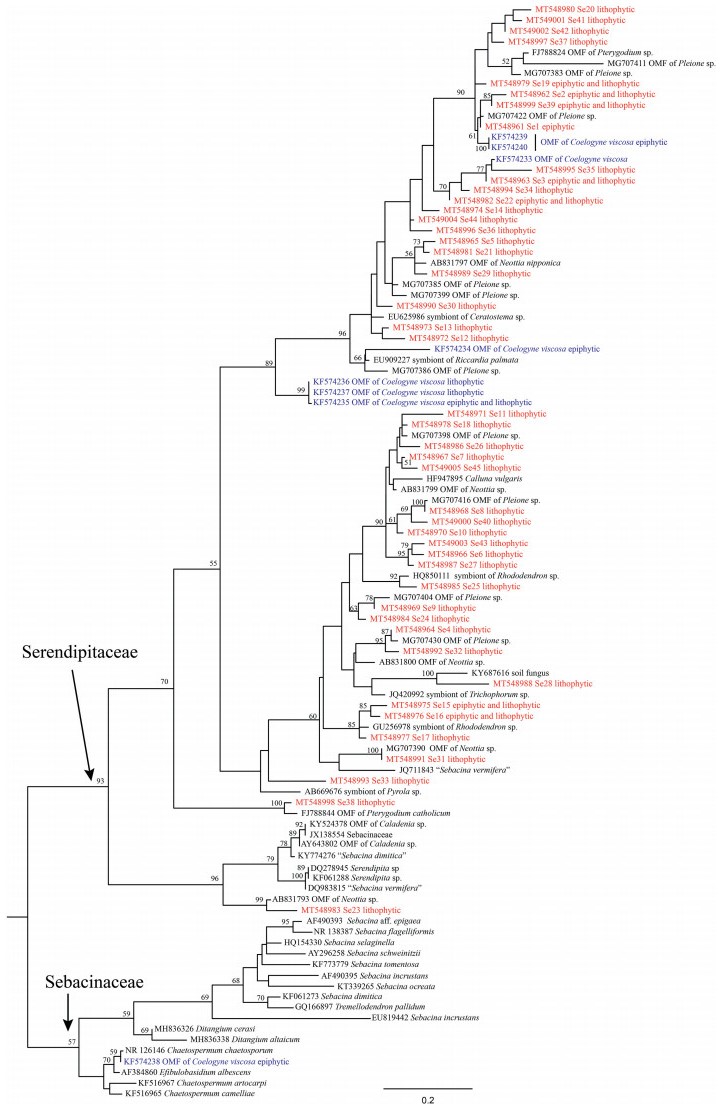
3. Results 3.1. Orchid mycorrhizal fungal OTUs and their phylogenetic relationshipsFrom the mycorrhizae of C. corymbosa, fungal sequences of the ITS region were clustered as 670 and 1984 OTUs for ITS3 and ITS86F sequencing, respectively. Among these OTUs, 559 for ITS3 sequencing and 1861 for ITS86F were included in the homogenized sequence abundance table, 58 for ITS3 and 46 for ITS86F were then designated as putative OMF and eventually merged into 73 OTUs (including 27 OTUs specific to ITS3, 15 specific to ITS86F, and 31 shared by both). To show the phylogenetic affiliations of the OMF, the OTUs summarized from two primer pairs were relabeled as Ce1-15, Se1-45, Th1-8, and Tu1-5, representing OMF belonging to Ceratobasidiaceae (15), Serendipitaceae (45), Thelephoraceae (8), and Tulasnellaceae (5), respectively (Supplementary Table 1). The OTUs of Tulasnellaceae and Ceratobasidiaceae from C. corymbosa were clustered into several phylogenetic clades, which showed divergence from those of C. viscosa (Fig. 2). The OTUs of Sebacinales from species of Coelogyne were mostly clustered into Serendipitaceae, except that one fungal sequence of C. viscosa was grouped in Sebacinaceae (Fig. 3).
|
| Fig. 2 Phylogenetic tree of Cantharellales fungi inferred from sequences of ITS2 region. Bootstrap values (> 50) are shown along nodes. Mycobionts detected from Coelogyne corymbosa and other species are indicated by red and blue, respectively. Detailed information on the fungal OTUs detected in this study is shown in Supplementary Table 1. |
|
| Fig. 3 Phylogenetic tree of Sebacinales fungi inferred from sequences of ITS2 region. Bootstrap values (> 50) are shown along nodes. Mycobionts detected from Coelogyne corymbosa and other species are indicated by red and blue, respectively. Detailed information on the fungal OTUs detected in this study is shown in Supplementary Table 1. |
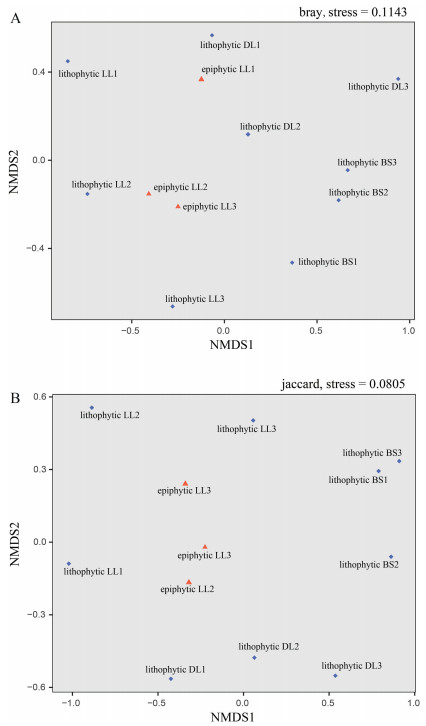
The OMF OTU abundance and presence data of ITS3 and ITS86F sequencing were merged and those of 61 OTUs (including 20 OTUs specific to ITS3 sequencing, 10 specific to ITS86F, and 31 shared) were eventually used in the NMDS and Permanova analyses, whereas data of the OTUs with less than five sequences (Supplementary Table 1) were excluded, as recommended by previous studies (Lindahl et al., 2013; Wang et al., 2019). The NMDS plots of the OMF communities detected from the lithophytes were not concentrated but intermixed by the plots of epiphytes in Fig. 4. The results of Permanova analyses showed that the variations among fungal communities of different life forms and sites were mostly not significant (Table 1).
|
| Fig. 4 Results of the NMDS analyses for OMF communities of each sample. A were generated based on the merged OTU abundance data of two primer pairs, B were based on binary data. Plots for mycobionts of lithophytic and epiphytic plants are indicated by blue diamonds and red triangles. LL, Longling; BS, Baoshan; and DL, Dali. |
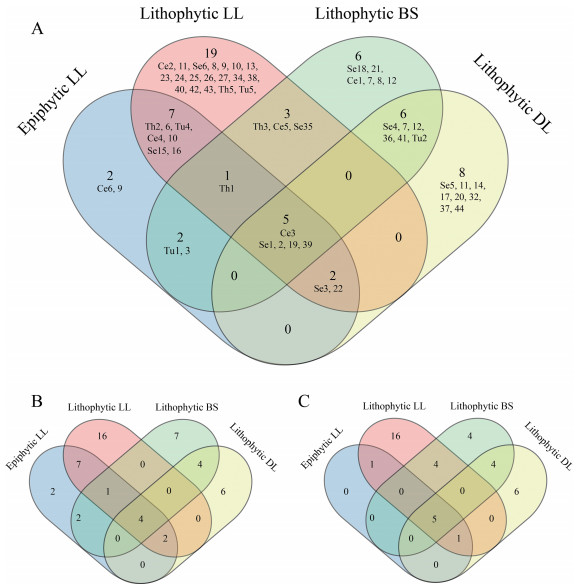
The OMF OTUs with less than five sequences were also not included in Venn diagrams. For the merged data of ITS3 and ITS86F sequencing, 17 OTUs were shared by epiphytic and lithophytic samples, which was up to 90% of the OTUs of epiphytic samples (Fig. 5A). Forty-two OTUs were specific to lithophytic orchids, of which 30 were identified as fungi of Serendipitaceae, six were from Ceratobasidiaceae, two were from Tulasnellaceae, and two were from Thelephoraceae (Fig. 5A). Fungi of Serendipitaceae also contributed a relatively large portion of the OTU communities specific to lithophytic orchids of two sites (15/19 for LL, and 8/8 for DL, respectively). Two other diagrams based on data of ITS3 and ITS86F sequencing were also provided (Fig. 5B–C), which showed that 16 and seven OTUs were shared by epiphytic and lithophytic individuals of C. corymbosa. Fourteen OTUs were shared by epiphytic and lithophytic samples of Longling (LL), based on ITS3 sequencing, while seven were shared based on ITS86F sequencing. This accounts for 78% (14/18) and 100% (7/7) of all OMF OTUs of the epiphytes (Fig. 5B–C).
|
| Fig. 5 Venn diagrams of OMF OTUs shared by different populations. A. Summarized from the combined data. B. Detected from ITS3 sequences. C. From ITS86F sequences. LL, Longling; BS, Baoshan; DL, Dali; Se, OTUs of Serendipitaceae; Ce, Ceratobasidiaceae; Tu, Tulasnellaceae; and Th, Thelephoraceae. |
Only two OTUs of Ceratobasidiaceae were reported from C. viscosa (Xing et al., 2015), and 15 were detected from C. corymbosa (Fig. 2; Supplementary Table 1). A similar situation was found in Serendipitaceae, which contributed the greatest number of OTUs for C. corymbosa. Seven sequences (KF574233-KF574237, and KF574239-KF574240) generated from C. viscosa (Xing et al., 2015) were grouped in four terminal nodes in our phylogeny of Serendipitaceae (Fig. 3). However, the OTUs of C. corymbosa grouped in 45 terminal nodes of Serendipitaceae, consisting of 12 specific to ITS3, 11 specific to ITS86F, and 22 shared by both (Fig. 3; Supplementary Table 1). This might be caused by OMF differences of each orchid species, but also might be due to the small data volume of Sanger sequencing on C. viscosa, as well as the primer pairs chosen. In previous fungal studies of C. ovalis and Coelogyne nervosa, no member of Ceratobasidiaceae or Sebacinales was found (Sathiyadash et al., 2014; Xing et al., 2019). In Tulasnellaceae, nine fungal OTUs of C. viscosa and two of C. ovalis were detected as OMF, and one was reported in a germination experiment of C. nervosa (Sathiyadash et al., 2014; Xing et al., 2015, 2019). In our study, five OTUs of Tulasnellaceae were detected from the mycorrhizae of C. corymbosa (Fig. 2; Supplementary Table 1).
Regardless of the number of OTUs detected, no fungal OTU from Cantharellales and Sebacinales was shared by C. corymbosa and the other three species of Coelogyne (viz., C. viscosa, C. nervosa, and C. ovalis) (Fig. 2, Fig. 3). At the fungal family level, although Tulasnellaceae, Ceratobasidiaceae, and Serendipitaceae, were all detected from the mycorrhizae of C. corymbosa and C. viscosa, fungi of Sebacinaceae were only found for the latter species (Fig. 2, Fig. 3; Xing et al., 2015). These findings strongly support the notion that orchid species prefer specific fungi. Alternatively, the absence of Sebacinaceae fungi for C. viscosa in our study may be explained by primer choice. We noticed that C. ovalis and C. viscosa shared two OTUs of Tulasnellaceae (MH005846 and MH005918 in Fig. 2, also see Xing et al., 2019). The fungal similarity between the two tropical orchids and the unique OMF OTUs of the subalpine C. corymbosa may be partially explained by geographical differences, although sampling of more species from a wider geographic distribution is still needed. In previous studies, arbuscular mycorrhizal, ectomycorrhizal, and ericoid mycorrhizal fungal communities have been found to be shaped by elevational gradients (Gai et al., 2012; Gorzelak et al., 2012; Looney et al., 2016). Mycorrhizal fungi of one terrestrial orchid species also change with latitudinal variation (Duffy et al., 2019), and further comparisons of the mycobionts of epiphytic orchids along elevational and latitudinal gradients will be meaningful.
4.2. The OMF communities of C. corymbosa were not affected by life form (lithophytic vs. epiphytic)No significant correlation between the OMF composition, the life form, and the collecting site was found, according to the results of the NMDS and Permanova analyses based on the merged data of two primer pairs. In the Venn diagrams and phylogenetic trees, the OMF OTUs of epiphytes were mostly shared by lithophytes, despite lithophytes harboring many specific OTUs. The mycorrhizae of the epiphytes and lithophytes of C. corymbosa were both covered by thick bryophytes, which might provide similar water and nutrient supplies, and partially explain the common fungal OTUs in mycorrhizae from both the arboreal and adjacent rocky environments.
A previous study found that the OMFs of C. viscosa are affected by life form; specifically, more OTUs were detected in epiphytic than in lithophytic plants (Xing et al., 2015). Aside from geographic distribution and species-specific differences, this variation may be partially explained by the relatively extreme environments of C. viscosa lithophytes, which live on rocky surfaces without bryophytes (Fig. 1b of Xing et al., 2015).
Pleione albiflora Cribb & C. Z. Tang were sampled at the same sites as those of C. corymbosa. Although OMF detection was based on sequencing and less data were generated for this species, the OMF OTUs of epiphytes in P. albiflora were also mostly shared by lithophytes (Supplementary Fig. 1; Qin et al., 2019). The mycobionts of lithophytic plants were more diverse on the whole but less diverse in the Longling population of P. albiflora, while the OTUs of lithophytes in all the sites were more diverse than those of epiphytes in C. corymbosa. The different OMF composition characters of the two orchids might also be related to their ecophysiological differences. C. corymbosa has evergreen leaves and persistent roots that are covered with a velamen consisting of four layers of dead cells. However, P. albiflora are characterized by annually renewed leaves and roots, and the absence of a velamen radicum (Zhang et al., 2016).
On the other hand, the unique fungal OTUs of lithophytes and epiphytes of C. viscosa and P. albiflora largely belong to Serendipitaceae, not Tulasnellaceae (Xing et al., 2015; Qin et al., 2019). This is consistent with the results of C. corymbosa in our study (Fig. 5 and Supplementary Fig. 1), although the fungal data for C. viscosa and P. albiflora were generated by using other primers in Sanger sequencing. In C. corymbosa, the OTUs specific to lithophytes mostly belonged to Serendipitaceae, and most fungi of this lineage were also specific to each lithophytic population (from Longling and Dali; Fig. 5A). Why were the Sebacinales fungal communities significantly different in the two growth habitats, but the Tulasnellaceae fungi not? According to report of Weiβ et al. (2011), the Sebacinales are not only extremely versatile in mycorrhizal associations but also present as universal symptomless endophytes. Given that most members of this fungal lineage are still un-culturable, it is still hard to answer if the differences among lithophytic and epiphytic habitats are caused by the endophytic habit of Sebacinales. To further corroborate OMF composition of epiphytic and lithophytic orchids in different elevational distributions, molecular data based on more extensive sampling are still needed. Overall, these findings improve our understandings of the OMF of different orchid life forms and provide novel insights for future research into the cultivation of Coelogyne.
Author contributionJ-H Wang and J Qin conceived and designated the experiments. J Qin and W Zhang collected and analyzed the data. J Qin wrote the initial manuscript, S-B Zhang and W Zhang revised the manuscript.
Declaration of Competing InterestOn behalf of the co-authors, I declare that the work is original, and has neither been published previously, nor is under consideration for publication elsewhere, in whole or in part. No conflict of interest exits in the submission of this manuscript, and the manuscript has been approved by all co-authors.
AcknowledgementsWe thank Jia-Lin Huang and Jia-Wei Li for their kind help with field work. John Meadows and Raymond Porter are acknowledged for improvements of the manuscript. This study was supported by the National Natural Science Foundation of China (Nos. 31670342 and 31700026), Yunnan Applied Basic Research Project (2019FB019), the Science and Technology Plan of Yunnan (2018BB010) and the Scientific and Technological Leading Talent Project of Yunnan Province (2016HA005).
Appendix A. Supplementary dataSupplementary data to this article can be found online at https://doi.org/10.1016/j.pld.2020.07.005.
Anderson, M.J., Gorley, R.N., Clarke, K.R., 2008. PERMANOVA+ for PRIMER: Guide to Software and Statistical Methods. PRIMER-E, Plymouth.
|
Bengtsson-Palme J., Ryberg M., Hartmann M., et al, 2013. Improved software detection and extraction of ITS1 and ITS2 from ribosomal ITS sequences of fungi and other eukaryotes for analysis of environmental sequencing data. Methods Ecol. Evol, 4: 914-919. DOI:10.1111/2041-210X.12073 |
Caporaso J.G., Kuczynski J., Stombaugh J., et al, 2010. QⅡME allows analysis of high-throughput community sequencing data. Nat. Methods, 7: 335-336. DOI:10.1038/nmeth.f.303 |
Chen, X., Liu, Z.J., Zhu, G.H., et al., 2009. Vol. 25 Orchidaceae. In: Wu, Z.Y., Raven, P.H., Hong, D.Y. (Eds.), Flora of China. Missouri Botanical Garden Press and Science Press, St. Louis and Beijing, pp. 315-325.
|
Dearnaley, J., Martos, F., Selosse, M.-A., 2012. Orchid mycorrhizas: molecular ecology, physiology, evolution and conservation aspects. In: Esser, K. (Ed.), The Mycota Volume IX-Fungal Associations, second ed. Springer-Verlag, Berlin, pp. 207-230.
|
Duffy K.J., Waud M., Schatz B., et al, 2019. Latitudinal variation in mycorrhizal diversity associated with a European orchid. J. Biogeogr, 46: 968-980. DOI:10.1111/jbi.13548 |
Edgar R.C., 2013. UPARSE: highly accurate OTU sequences from microbial amplicon reads. Nat. Methods, 10: 996-998. DOI:10.1038/Nmeth.2604 |
Edgar R.C., Flyvbjerg H., 2015. Error filtering, pair assembly and error correction for next-generation sequencing reads. Bioinformatics, 31: 3476-3482. DOI:10.1093/bioinformatics/btv401 |
Gai J.P., Tian H., Yang F.Y., et al, 2012. Arbuscular mycorrhizal fungal diversity along a Tibetan elevation gradient. Pedobiologia, 55: 145-151. DOI:10.1016/j.pedobi.2011.12.004 |
Gardes M., White T.J., Fortin J.A., et al, 1991. Identification of indigenous and introduced symbiotic fungi in ectomycorrhizae by amplification of nuclear and mitochondrial ribosomal DNA. Can. J. Bot, 69: 180-190. DOI:10.1139/b91-026 |
Gorzelak M.A., Hambleton S., Massicotte H.B., 2012. Community structure of ericoid mycorrhizas and root-associated fungi of Vaccinium membranaceum across an elevation gradient in the Canadian Rocky Mountains. Fungal Ecol, 5: 36-45. DOI:10.1016/j.funeco.2011.08.008 |
Gravendeel B., Chase M.W., De Vogel E.F., et al, 2001. Molecular phylogeny of Coelogyne (Epidendroideae; Orchidaceae) based on plastid RFLPS, matK, and nuclear ribosomal its sequences: evidence for polyphyly. Am. J. Bot, 88: 1915-1927. DOI:10.2307/3558367 |
Gravendeel B., Smithson A., Slik F.J.W., et al, 2004. Epiphytism and pollinator specialization: drivers for orchid diversity?. Philos. Trans. R. Soc. Lond. Ser. B Biol. Sci, 359: 1523-1535. DOI:10.1098/rstb.2004.1529 |
Hall T.A., 1999. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser, 41: 95-98. |
Jacquemyn H., Waud M., Brys R., et al, 2017. Mycorrhizal associations and trophic modes in coexisting Orchids: an ecological continuum between auto- and mixotrophy. Front. Plant Sci, 8. DOI:10.3389/fpls.2017.01497 |
Katoh K., Standley D.M., 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol, 30: 772-780. DOI:10.1093/molbev/mst010 |
Kõljalg U., Larsson K.H., Abarenkov K., et al, 2005. UNITE: a database providing web-based methods for the molecular identification of ectomycorrhizal fungi. New Phytol, 166: 1063-1068. DOI:10.1111/j.1469-8137.2005.01376.x |
Lindahl B.D., Nilsson R.H., Tedersoo L., et al, 2013. Fungal community analysis by high-throughput sequencing of amplified markers - a user's guide. New Phytol, 199: 288-299. DOI:10.1111/nph.12243 |
Looney B.P., Ryberg M., Hampe F., et al, 2016. Into and out of the tropics: global diversification patterns in a hyperdiverse clade of ectomycorrhizal fungi. Mol.Ecol, 25: 630-647. DOI:10.1111/mec.13506 |
Martin M., 2011. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet. J, 17: 10-12. DOI:10.14806/ej.17.1.200 |
Martos F., Munoz F., Pailler T., et al, 2012. The role of epiphytism in architecture and evolutionary constraint within mycorrhizal networks of tropical orchids. Mol. Ecol, 21: 5098-5109. DOI:10.1111/j.1365-294X.2012.05692.x |
Nylander, J., 2004. MrModeltest 2.2. Computer Software Distributed by the University of Uppsala, Sweden. Evolutionary Biology Centre. Uppsala University, Uppsala.
|
Oja J., Kohout P., Tedersoo L., et al, 2015. Temporal patterns of orchid mycorrhizal fungi in meadows and forests as revealed by 454 pyrosequencing. New Phytol, 205: 1608-1618. DOI:10.1111/nph.13223 |
Oksanen, J., Blanchet, F.G., Kindt, R., et al., 2013. Vegan: community ecology package. R Package Version 2.0-0. http://CRAN.R-project.org/package=vegan.
|
Qin J., Zhang W., Ge Z.-W., et al, 2019. Molecular identifications uncover diverse fungal symbionts of Pleione (Orchidaceae). Fungal Ecol, 37: 19-29. DOI:10.1016/j.funeco.2018.10.003 |
Sathiyadash K., Muthukumar T., Murugan S.B., et al, 2014. In vitro symbiotic seed germination of South Indian endemic orchid Coelogyne nervosa. Mycoscience, 55: 183-189. DOI:10.1016/j.myc.2013.08.005 |
Stamatakis A., 2006. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics, 22: 2688-2690. DOI:10.1093/bioinformatics/btl446 |
Taylor D.L., McCormick M.K., 2008. Internal transcribed spacer primers and sequences for improved characterization of basidiomycetous orchid mycorrhizas. New Phytol, 177: 1020-1033. DOI:10.1111/j.1469-8137.2007.02320.x |
Těšitelová T., Kotilinek M., Jersakova J., et al, 2015. Two widespread green Neottia species (Orchidaceae) show mycorrhizal preference for Sebacinales in various habitats and ontogenetic stages. Mol. Ecol, 24: 1122-1134. DOI:10.1111/mec.13088 |
Turenne C.Y., Sanche S.E., Hoban D.J., et al, 2000. Rapid identification of fungi by using the ITS2 genetic region and an automated fluorescent capillary electrophoresis system (vol 37, p 1846, 1999). J. Clin. Microbiol, 38: 944-944. DOI:10.1128/JCM.38.2.944-944.2000 |
Wang Z.H., Jiang Y., Deane D.C., et al, 2019. Effects of host phylogeny, habitat and spatial proximity on host specificity and diversity of pathogenic and mycorrhizal fungi in a subtropical forest. New Phytol, 223: 462-474. DOI:10.1111/nph.15786 |
Waud M., Busschaert P., Lievens B., et al, 2016. Specificity and localised distribution of mycorrhizal fungi in the soil may contribute to co-existence of orchid species. Fungal Ecol, 20: 155-165. DOI:10.1016/j.funeco.2015.12.008 |
Waud M., Busschaert P., Ruyters S., et al, 2014. Impact of primer choice on characterization of orchid mycorrhizal communities using 454 pyrosequencing. Mol. Ecol. Resour, 14: 679-699. DOI:10.1111/1755-0998.12229 |
Weiß M., Sýkorová Z., Garnica S., et al, 2011. Sebacinales everywhere: previously overlooked ubiquitous fungal endophytes. PloS One, 6: e16793. DOI:10.1371/journal.pone.0016793 |
Xing X.-K., Gai X.-G., Liu Q., et al, 2015. Mycorrhizal fungal diversity and community composition in a lithophytic and epiphytic orchid. Mycorrhiza, 25: 289-296. DOI:10.1007/s00572-014-0612-5 |
Xing X.-K., Jacquemyn H., Gai X.-G., et al, 2019. The impact of life form on the architecture of orchid mycorrhizal networks in tropical forest. Oikos, 128: 1254-1264. DOI:10.1111/oik.06363 |
Yukawa T., Ogura-Tsujita Y., Shefferson R.P., et al, 2009. Mycorrhizal diversity in Apostasia (Orchidaceae) indicates the origin and evolution of orchid mycorrhiza. Am. J. Bot, 96: 1997-2009. DOI:10.3732/ajb.0900101 |
Zhang S.-B., Yang Y.-J., Li J.-W., et al, 2018. Physiological diversity of orchids. Plant Divers, 40: 196-208. DOI:10.1016/j.pld.2018.06.003 |
Zhang W., Hu H., Zhang S.-B., 2016. Divergent adaptive strategies by two cooccurring epiphytic orchids to water stress: escape or avoidance?. Front. Plant Sci, 7: 588. DOI:10.3389/fpls.2016.00588 |



