b. Key Laboratory of Ecology of Rare and Endangered Species and Environmental Protection, Ministry of Education, Guangxi Normal University, Guilin, China;
c. Longtan Nature Reserve Management Center, Hechi, China;
1. College of Life Science, Guangxi Normal University, No. 1 Yanzhong Road, Guilin, Guangxi 541006, China;
2. Longtan Nature Reserve Management Center, Hechi, China
Conservation of rare and endangered species and their sustainable utilization relies on research into their genetic diversity, population differentiation, and gene flow (Yang et al., 2010, Zaya et al., 2017). Genetic diversity is an important part of conservation biodiversity (Ramanatha Rao and Hodgkin, 2002) and essential for maintaining both the ability to cope with environmental changes and the evolutionary potential of species (Bhattacharyya and Kumaria, 2015, Ellstrand and Elam, 1993). Genetic variation may be correlated with the geographical distribution range of species (Solórzano et al., 2016). Species with a wide geographical range might maintain higher levels of genetic diversity than species with limited range (Chung et al., 2018, Levy et al., 2016). In particular, rare or endemic species with restricted distribution range and small population size are characterized by poor genetics (Solórzano et al., 2016, Zaya et al., 2017).
Camellia huana T. L. Ming et W. J. Zhang is a small shrub with yellow flowers belonging to Camellia sect. Archecamellia Sealy (Theaceae) (Fig. 1). The distribution of C. huana is very narrow, being restricted to evergreen, broad-leaved forests in the limestone hills of Tian'e County in northern Guangxi and Luodian County, Ceheng County and Xingren County in southern Guizhou, China. C. huana is the most northerly species of sect. Archecamellia and shows a discontinuous distribution with other sect. Archecamellia species. It may be better adapted to northern climates (Logan et al., 2019) and is of great significance in the introduction of yellow-flowered Camellia to the north. Although the total number of C. huana plants was once more than 310, 000, by 2013, only about 30, 000 individuals existed in natural populations (An, 2005, Xie et al., 2014). A great number of C. huana plants have been transplanted because C. huana is an ornamental plant and its flowers and leaves can be used as tea. C. huana is listed as an Endangered (EN) species on the Threatened Species List of China's Higher Plants (Qin et al., 2017).

|
| Fig. 1 Camellia huana. Wild plant (a), flower (b), fruit (c). |
In the present study, we combined biparentally inherited microsatellites (simple sequence repeats; SSRs) with uniparentally inherited chloroplast DNA (cpDNA) to investigate the genetic diversity and structure of C. huana. Microsatellites are effective molecular tools for assessing genetic variation, characterized by their high variability, low cost of detection, large distribution and co-dominant inheritance (Abbasov et al., 2018, Spooner et al., 2007). cpDNA displays conserved gene order, low mutation rate and a lower effective population size than nuclear genes, and is thus suitable for providing supplementary and more accurate information on population variation and differentiation (Huang et al., 2011). In C. huana, cpDNA is maternally inherited and dispersed by seeds (Birky, 2008). This study aimed to (1) estimate the level of genetic diversity within and between C. huana populations, (2) infer the genetic structure and differentiation of C. huana, and (3) develop effective conservation strategies and management measures.
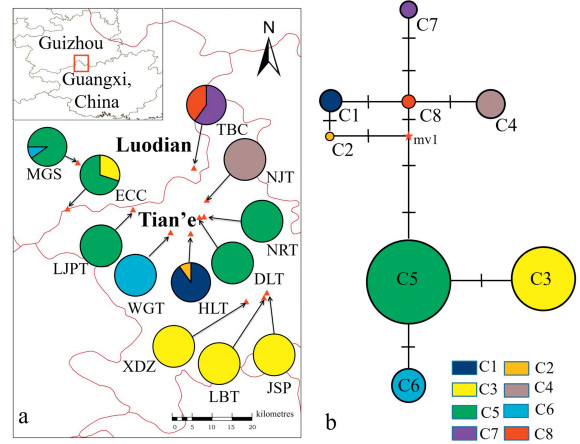
2. Materials and methods 2.1. Plant materialsThe entire distribution area of C. huana was surveyed in 2018 and 2019, and only five natural populations were found, in Tian'e County, Guangxi. Four populations were from the Longtan Nature Reserve (populations DLT, NRT, HLT and JSP). A total of 358 individual C. huana samples were collected from these five natural populations and seven other populations directly transplanted from wild habitats: three in the south of Guizhou province, and the remaining four in the north of Guangxi. Twenty-nine to thirty individuals were collected randomly from each population. One plant was sampled approximately every 10–50 m in the natural populations, depending on the size of each population. The location and number of sampled individuals for each population are provided in Table 1 and Fig. 2a.
| Population code | Population location | Longitude (E)/latitude (N) | Elevation (m) | Estimated census size | Sample size SSR (cpDNA) | cpDNA | SSR | ||||||||
| Haplotypes (number) | Haplotype diversity (Hd) | Nucleotide diversity (Pi) | NA | NE | HO | HE | F | PPB (%) | |||||||
| Maoguishan (MGS) population | Xiaogan village, Luokun town | 106°40'/25°17′ | 861 | – | 30 (10) | C5 (9) C6 (1) |
0.200 | 0.00004 | 5.417 | 2.908 | 0.506 | 0.605 | 0.161 | 100.00 | |
| Ecuncun (ECC) population | E village, Hongshuihe town | 106°40'/25°11′ | 394 | – | 30 (10) | C3 (3) C5 (7) |
0.467 | 0.00009 | 4.833 | 2.629 | 0.478 | 0.531 | 0.177 | 100.00 | |
| Tianbacun (TBC) population | Tianba village, Maojin town | 106°57'/25°16′ | 1048 | – | 30 (10) | C7 (6) C8 (4) |
0.533 | 0.00021 | 4.333 | 2.601 | 0.589 | 0.566 | -0.054 | 91.67 | |
| Laojiangpingtun (LJPT) population | Naming village, Xialao town |
106°50'/25°10′ | 734 | – | 30 (10) | C5 (10) | 0 | 0 | 3.917 | 2.279 | 0.378 | 0.446 | 0.196 | 100.00 | |
| Weiguantun (WGT) population | Niuchang village, Xiangyang town | 106°55'/25°07′ | 854 | – | 29 (10) | C6 (10) | 0 | 0 | 4.250 | 2.351 | 0.440 | 0.464 | 0.042 | 91.67 | |
| Najietun (NJTa) population | Hekou village, Pojie town | 107°00'/25°11′ | 840 | – | 30 (10) | C4 (10) | 0 | 0 | 4.333 | 2.230 | 0.433 | 0.455 | 0.009 | 91.67 | |
| Daliangtun (DLTa) population | Pojie village, Pojie town | 106°59'/25°09′ | 525 | 210 | 30 (10) | C5 (10) | 0 | 0 | 3.750 | 2.002 | 0.458 | 0.420 | -0.095 | 83.33 | |
| Narangtun (NRTa) population | Pojie village, Pojie town | 107°00'/25°09′ | 680 | 16011 | 30 (10) | C5 (10) | 0 | 0 | 3.417 | 2.410 | 0.542 | 0.515 | -0.065 | 100.00 | |
| Henglitun (HLTa) population | Quanping village, Xiangyang town | 106°58'/25°07′ | 508 | 600 | 30 (10) | C1 (9) C2 (1) |
0.200 | 0.00004 | 4.167 | 2.458 | 0.528 | 0.505 | -0.050 | 91.67 | |
| Xiaodongzi (XDZ) population | Long'e village, Bala town | 107°06'/24°58′ | 900 | – | 30 (10) | C3 (10) | 0 | 0 | 2.833 | 1.722 | 0.411 | 0.356 | -0.130 | 83.33 | |
| Jianshipo (JSPa) population | Tangying village, Liupai town | 107°09'/24°58′ | 732 | 200 | 30 (10) | C3 (10) | 0 | 0 | 3.083 | 1.729 | 0.353 | 0.339 | 0.038 | 91.67 | |
| Longbutun (LBT) population | Tangying village, Liupai town | 107°09'/24°57′ | 804 | – | 29 (10) | C3 (10) | 0 | 0 | 2.833 | 1.946 | 0.374 | 0.388 | 0.014 | 83.33 | |
| Total | 17021 | 358 (120) | 0.759 | 0.00042 | |||||||||||
| Mean | 3.931 | 2.272 | 0.457 | 0.466 | 0.025 | 92.36 | |||||||||
| NA, number of mean alleles; NE, number of effective alleles; HO, observed heterozygosity; HE, expected heterozygosity; F, fixation index; PPB, percentage of polymorphic loci. a Natural population; the remaining populations were transplanted directly from wild habitats. |
|||||||||||||||
|
| Fig. 2 Distribution of 12 Camellia huana populations (see Table 1 for population codes). Distribution (a) and network (b) of cpDNA haplotypes detected among 12 populations of C. huana. In (b), the size of the circle is proportional to the frequency of each sampled haplotype and the lines on the branches indicate the number of steps separating adjacent haplotypes. |
Healthy, fresh leaves were harvested and stored with silica gel in zip-lock plastic bags until DNA isolation. Genomic DNA was extracted according to a modified CTAB method (Doyle and Doyle, 1990). Extracted DNA was dissolved in 50 μL 1 × TE buffer for use as PCR templates. Primer pairs described by Wei et al. (2017) and Xi et al. (2012) were adopted for amplifying a cpDNA fragment from the small single-copy (SSC) region comprising the following three fragments: SSC1, CP30 and SSC3. PCR products were sequenced using the same primers plus four internal sequencing primers: SSC1-1, CP30-1, CP30-2 and SSC3-1 (Supplemental file 1: Table S1).
PCR amplification of cpDNA was carried out in 50 μL reactions containing 5 μL 10 × PCR buffer (Mg2+ plus), 4 μL dNTP mix (2.5 mM), 0.5 μL each primer (50 μM), 0.5 μL rTaq DNA Polymerase (all reagents from TaKaRa, China), 2.0 μL DNA (20 ng) and 37.5 μL double-distilled water. PCR amplification was performed in a thermocycler under the following conditions: initial 5 min denaturation at 94 ℃; followed by 35 cycles of 1 min at 94 ℃, 30 s annealing at specific annealing temperatures, 1 min elongation at 72 ℃; and final extension for 10 min at 72 ℃. All PCR products were directly purified and sequenced. All chloroplast haplotype sequences generated in this study were deposited in GenBank under accession numbers MN380425–MN380432.
Microsatellite markers were selected from previously developed microsatellites in Camellia pingguoensis D. Fang (Lu et al., 2014) and Camellia flavida H. T. Chang (Liufu et al., 2014). PCR amplification was carried out in 20 μL reactions containing 2 μL 10 × PCR buffer (Mg2+ plus), 0.4 μL dNTP mix (10 mM), 0.2 μL each primer (50 μM), 0.2 μL rTaq DNA Polymerase (all reagents from TaKaRa, China), 1.0 μL DNA (20 ng) and 16 μL double-distilled water. PCR amplifications were performed in a thermocycler under the same conditions as cpDNA fragment amplification. Preliminary screening identified 12 microsatellite loci (Supplemental file 1: Table S2) for further use in this study. The selected primer pairs were labeled with a fluorescent dye at the 5′ end, and their PCR products were separated and visualized using an ABI3730 automated sequencer (Applied Biosystems, USA). Microsatellite marker profiles were read using the Genemarker software (Softgenetics, USA).
2.3. cpDNA sequence analysisAll sequences were aligned using MUSCLE (Edgar, 2004). Population genetic parameters, including the number of haplotypes (n), haplotype diversity (Hd), nucleotide diversity (Pi) and number of polymorphic sites (S), were calculated for aligned cpDNA sequences using DNAsp version 5.0 (Librado and Rozas, 2009).
The software PermutCpSSR (Pons and Petit, 1996) was employed for evaluating differentiation for unordered alleles (GST) and ordered alleles (NST) under 1000 stochastic permutations, to test for the presence of phylogeographic structures with significant differences between GST and NST. Analysis of molecular variance (AMOVA) (Excoffier et al., 1992) was implemented using Arlequin 3.0 (https://www.fousoft.com/arlequin.html), to study the genetic variation partitioned within and among populations. Meanwhile, pairwise fixation indices (FST) were also calculated using this method. Any correlation between the matrices of FST genetic differentiation and geographical distance were studied by performing a Mantel test (Mantel, 1967) using GenAIEx6.5 (http://biology-assets.anu.edu.au/GenAlEx/Welcome.html).
Network analysis (http://www.fluxus-engineering.com/sharenet.htm) was performed to construct a haplotype network using the median-joining method (Bandelt et al., 1999), analyzing the genealogical relationships among all the haplotypes. ArcMap GIS (http://desktop.arcgis.com) was then used to map the geographical distribution of populations and haplotypes.
2.4. SSR analysisGenAIEx6.5 (http://biology-assets.anu.edu.au/GenAlEx/Welcome.html) was used for dataset editing and formatting. Genepop version 4.2.2 (http://genepop.curtin.edu.au/) was used for testing departure from Hardy–Weinberg equilibrium and estimating linkage disequilibrium, and the sequential Bonferroni method (Rice, 1989) was used to correct significance levels for each population and each locus. Differentiation between pairwise populations based on FST was calculated using Arlequin 3.0 (https://www.fousoft.com/arlequin.html), and gene flow between population pairs was estimated using Wright's method, Nm = (1-FST)/4FST (Wright, 1931). Genetic parameters, consisting of mean number of alleles (NA), observed heterozygosity (Ho), expected heterozygosity (HE), percentage of polymorphic loci (PPB), fixation index (F) and inbreeding coefficient (Fis) for each microsatellite locus, were calculated using GenAIEx6.5.
An admixture model based on the Bayesian clustering method was performed with SSR data in STRUCTURE 2.3 (http://web.stanford.edu/group/pritchardlab/structure.html) to determine whether species were genetically distinct. A total of 20 independent simulations were run for a K-value of 1-13 with 100, 000 burn-in steps followed by 500, 000 steps. Structure harvester (http://taylor0.biology.ucla.edu/structureHarvester/) was then utilized to estimate the most likely number (K) (Rosenberg et al., 2001) of genetic clusters based on both log-likelihood L (K) and delta K (ΔK) (Evanno et al., 2005) methods. Principal coordinate analysis (PCoA) was carried out using GenAIEx6.5. Mantel tests in GenAIEx6.5 were performed to estimate isolation by distance for SSR data, using a correlation of FST/(1-FST) (Mantel, 1967) with geographic distance for all pairs of populations.
Bottleneck 1.2.02 (Cornuet and Luikart, 1996) was used to test for recent bottleneck events in the species. The demographic history of populations was explored under the stepwise mutation model (SMM) and two-phased model (TPM), and by a heterozygosity excess test. Computation was performed using two methods (Sign and Wilcoxon tests) appropriate for a population number < 20.
3. Results 3.1. cpDNA sequencescpDNA consensus sequences from 120 individuals (10 per population) were 5200 bp in length containing nine polymorphic sites with no indels (Table 2). We identified eight chloroplast haplotypes (Table 1). Haplotypes C1 and C2, and haplotypes C7 and C8 were unique to populations HLT and TBC, respectively; haplotype C4 was fixed for population NJT. Haplotype C5 was the most abundant haplotype with wide distribution in five populations (MGS, ECC, LJPT, DLT and NRT); haplotypes C3 and C6 were shared by four populations (ECC, XDZ, JSP and LBT) and two populations (MGS and WGT), respectively. Genetic diversity indices of total nucleotide (Pi) and haplotype (Hd) diversity in all populations were 0.00042 (individually ranging from 0 to 0.00021) and 0.759 (individually ranging from 0 to 0.533), respectively, as inferred from cpDNA (Table 1). We detected cpDNA sequence variation only within four populations (MGS, ECC, TBC and HLT).
| Haplotype | Polymorphic sites | ||||||||
| 60 | 178 | 658 | 1425 | 1572 | 2610 | 2757 | 3285 | 5153 | |
| C1 | T | C | T | C | G | T | T | T | C |
| C2 | . | . | G | . | . | . | . | . | . |
| C3 | . | . | G | A | . | G | C | G | . |
| C4 | . | . | . | . | . | G | . | . | T |
| C5 | . | . | G | . | . | G | C | G | . |
| C6 | . | . | G | . | A | G | C | G | . |
| C7 | C | T | . | . | . | G | . | . | . |
| C8 | . | . | . | . | . | G | . | . | . |
To further study population genetic structures of C. huana, we examined genetic differentiation by detecting sequence variation in cpDNA sequences across the 12 natural and transplanted populations. Analysis of molecular variance (AMOVA) revealed that the majority of genetic variation (93.18%) existed between populations, while only 6.82% was partitioned within populations at the cpDNA level (Table 3). The FST value of 0.9318 also indicated relatively large differentiation between the sampled populations. In addition, no significant correlation was detected between genetic and geographic distance at the cpDNA level (R2 = 0.0189, P = 0.180) (Supplemental file 1: Fig. S1a).
| Marker | Source of variation | d.f. | Sum of squares | Variance components | Percentage of variation (%) |
| cpDNA | Among populations | 11 | 121.942 | 1.10051 | 93.18 |
| Within populations | 108 | 8.700 | 0.08056 | 6.82 | |
| SSR | Among populations | 11 | 545.180 | 0.78229 | 21.59 |
| Within populations | 704 | 2002.351 | 2.84425 | 78.41 | |
| d.f., Degrees of freedom. | |||||
To further investigate the relationship between haplotypes, we constructed a network using the median-joining method for cpDNA (Fig. 2b). Haplotypes C5 and C8 took central positions while the other haplotypes were distributed on outside nodes in the network. This distribution pattern suggested that haplotypes C5 and C8 likely represent the ancient haplotypes for this distribution area. We found a significantly larger NST of 0.932 than GST of 0.856 (P < 0.05) for cpDNA data, suggesting pairs of different cpDNA haplotypes from the same populations have more similar sequences than pairs from different populations.
3.2. SSR dataHardy–Weinberg equilibrium test results showed that four of the 144 locus–population combinations deviated from expectation after Bonferroni correction, and no significant linkage disequilibrium was found, suggesting that the 12 microsatellite loci represented independent information across all samples.
Diversity estimates were summarized for each locus (Supplemental file 1: Table S3). A total of 99 alleles at 12 SSR loci were revealed across 358 individuals (approximately 30 per population) from 12 C. huana populations. The total number of alleles (NT) per locus ranged from three (at locus FLA14) to 12 (at loci FLA11 and TER14), with a mean of 8.25 alleles per locus. The number of effective alleles (NE) per locus varied from 1.135 (at locus FLA14) to 3.430 (at locus TER2), with a mean of 2.272. The observed (Ho) and expected (HE) heterozygosity ranged from 0.053 (at locus FLA14) to 0.857 (at locus FLA21) and from 0.078 (at locus FLA14) to 0.656 (at locus FLA11), respectively.
Intra-population microsatellite variation parameters are listed in Table 1. The number of mean alleles (NA) per population varied from 2.833 (populations XDZ and LBT) to 5.417 (population MGS), with an average number of 3.931 across all populations. The average number of effective alleles (NE) for all populations was 2.272, varying from 1.722 (population XDZ) to 2.908 (population MGS). The average values of Ho and HE for all populations were 0.457 and 0.466, respectively; Ho values varied from 0.353 (population JSP) to 0.589 (population TBC) and HE values from 0.339 (population JSP) to 0.605 (population MGS). The fixation index (F) varied from -0.130 (population XDZ) to 0.196 (population LJPT), with a mean value of 0.025. The above parameters show that population MGS possesses the highest genetic diversity.
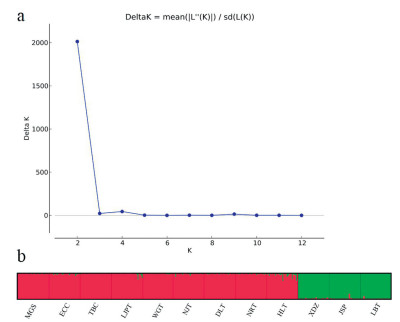
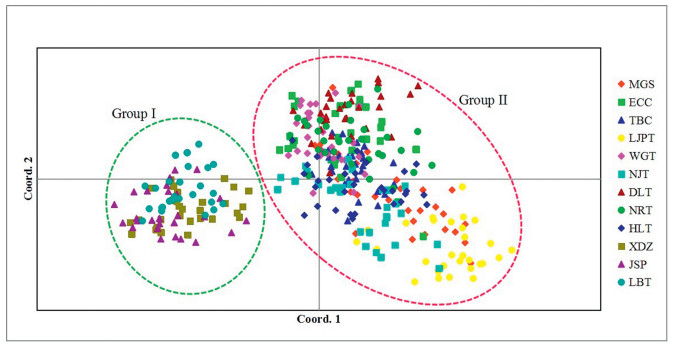
By STRUCTURE analysis of SSR data, we estimated the optimum number of groups (K) according to the procedure described by Evanno et al. (2005). The highest delta K (ΔK) for 358 individuals was K = 2 (Fig. 3a), suggesting that the 12 populations can be split into two groups. Populations XDZ, JSP and LBT were assigned to Group I; the remaining populations (MGS, ECC, TBC, LJPT, WGT, NJT, DLT, NRT and HLT) composed Group II (Fig. 3b). The existence of two groups was also supported by principal coordinate analysis (PCoA) (Fig. 4). cpDNA-based haplotype geographical distribution revealed that there was only one haplotype (C3) in Group I (populations XDZ, JSP and LBT; Fig. 2). Although haplotype C3 also existed in a cultivated population (ECC) of Group II, it may have been introduced from Group I; therefore, haplotype C3 may be a unique haplotype of group I.
|
| Fig. 3 Relationships between the number of genetic groups (K) and the estimated value delta K (ΔK) based on the SSR dataset (a). Estimated genetic clustering (K = 2) obtained with the STRUCTURE program for 12 populations of Camellia huana; each color represents a different cluster (b). |
|
| Fig. 4 Principal coordinate analysis (PCoA) of SSR phenotypes from 12 populations and 358 individuals of Camellia huana. |
AMOVA analysis of SSR data showed that 21.59% genetic variation resided between populations, and 78.41% within populations. Estimates of gene flow for each pair between the 12 populations are presented in Table 4. The maximum gene flow was observed between populations DLT and ECC (9.184), whereas the minimum gene flow was observed between populations LJPT and JSP (0.347). No significant effect of isolation by distance was discovered (Supplemental file 1: Fig. S1b) as the correlation between genetic and geographic distance was insignificant (R2 = 0.0234, P = 0.360), which was supported by the results of the Mantel test.
| MGS | ECC | TBC | LJPT | WGT | NJT | DLT | NRT | HLT | XDZ | JSP | LBT | |
| MGS | – | 3.208 | 3.138 | 1.852 | 1.636 | 3.324 | 1.419 | 1.591 | 2.336 | 0.707 | 0.658 | 0.744 |
| ECC | 0.072 | – | 2.518 | 0.807 | 3.084 | 1.590 | 9.184 | 2.290 | 1.527 | 0.800 | 0.694 | 0.920 |
| TBC | 0.074 | 0.090 | – | 0.830 | 1.796 | 1.857 | 1.159 | 1.290 | 1.562 | 0.665 | 0.617 | 0.843 |
| LJPT | 0.119 | 0.236 | 0.232 | – | 0.579 | 1.048 | 0.537 | 0.650 | 0.737 | 0.375 | 0.347 | 0.364 |
| WGT | 0.133 | 0.075 | 0.122 | 0.301 | – | 1.647 | 1.315 | 1.416 | 1.230 | 0.685 | 0.624 | 0.729 |
| NJT | 0.070 | 0.136 | 0.119 | 0.193 | 0.132 | – | 0.836 | 1.242 | 1.464 | 0.670 | 0.629 | 0.640 |
| DLT | 0.150 | 0.027 | 0.177 | 0.318 | 0.160 | 0.230 | – | 1.263 | 0.808 | 0.519 | 0.449 | 0.579 |
| NRT | 0.136 | 0.098 | 0.162 | 0.278 | 0.150 | 0.168 | 0.165 | – | 1.338 | 0.586 | 0.559 | 0.630 |
| HLT | 0.097 | 0.141 | 0.138 | 0.253 | 0.169 | 0.146 | 0.236 | 0.157 | – | 0.633 | 0.582 | 0.660 |
| XDZ | 0.261 | 0.238 | 0.273 | 0.400 | 0.268 | 0.272 | 0.325 | 0.299 | 0.283 | – | 1.489 | 1.040 |
| JSP | 0.275 | 0.265 | 0.288 | 0.419 | 0.286 | 0.284 | 0.357 | 0.309 | 0.300 | 0.144 | – | 1.897 |
| LBT | 0.251 | 0.265 | 0.229 | 0.407 | 0.255 | 0.281 | 0.302 | 0.284 | 0.275 | 0.194 | 0.116 | – |
Mutation-drift equilibrium estimated under different models and methods was calculated using Bottleneck analysis (Supplemental file 1: Table S4). This revealed that C. huana did not experience a population contraction. Under the TPM, populations TBC and NRT showed a significant excess of heterozygosity using the Wilcoxon test (P < 0.05); under the SMM, four populations (TBC, NJT, DLT and JSP) had a significant excess of heterozygosity using the Sign test. This indicates five populations (TBC, NJT, DLT, NRT and JSP) deviated from mutation–drift equilibrium. However, all populations had normal L-shaped distributions under the Mode shift model, suggesting that C. huana has not undergone a recent severe bottleneck.
4. Discussion 4.1. Chloroplast and microsatellite diversity in Camellia huanacpDNA and microsatellite markers in C. huana revealed a low level of genetic diversity (n = 8, Hd = 0.759, Pi = 0.00042 for cpDNA, NA = 3.931, HE = 0.466 for SSRs). Relationships between geographic range and level of genetic diversity, at both the population and species levels, tend to be strong (Solórzano et al., 2016). Theoretically, plant species with geographically restricted distribution often possess reduced genetic diversity compared with more widespread species (Coates et al., 2003, Yoichi and Tomaru, 2014, Levy et al., 2016, Chung et al., 2018). As expected, C. huana, with limited distribution, possessed relatively lower genetic diversity than many of its relatives with more widespread distribution based on similar markers, e.g., Camellia taliensis (W. W. Smith) Melchior (Liu et al., 2012, Zhao et al., 2014), C. flavida (Wei et al., 2017), C. sinensis (L.) O. Kuntze (Yao et al., 2012) and C. japonica L. (Ueno et al., 2000) (Table 5). Furthermore, the expected heterozygosity in C. huana measured by SSR markers (HE = 0.466) was slightly higher than the average value based on SSRs in endemic species (HE = 0.420) but considerably lower than that in regional or widespread species (HE = 0.650 and HE = 0.620) (Nybom, 2004). C. huana shows a discontinuous distribution with other yellow-flowered Camellia species, resulting in limited hybridization and introgression between C. huana and its relatives. This discontinuous distribution may also contribute to the low genetic diversity.
| SSR | cpDNA | References | |||||
| Number of mean alleles | Expected heterozygosity | Number of haplotypes | Haplotype diversity | Nucleotide diversity | |||
| Camellia huana | 3.931 | 0.466 | 8 | 0.759 | 0.00042 | This study | |
| Camellia taliensis | 6.776 | 0.597 | 12 | 0.84129 | 0.00314 | Liu et al. (2012); Zhao et al. (2014) |
|
| Camellia flavida | – | – | 17 | 0.94101 | 0.00157 | Wei et al. (2017) | |
| Camellia sinensis | 4.3 | 0.640 | – | – | – | Yao et al. (2012) | |
| Camellia japonica | 16.5 | 0.840 | – | – | – | Ueno et al. (2000) | |
| Camellia chrysanthoides | 3.7 | 0.431 | – | – | – | Chen et al. (2019) | |
| Camellia micrantha | 3.8 | 0.476 | – | – | – | Chen et al. (2019) | |
| Camellia parvipetala | 3.9 | 0.501 | – | – | – | Chen et al. (2019) | |
| Camellia nitidissima | 3.881 | 0.546 | 7 | 0.837 | 0.00082 | Li et al. (2019); Lu et al. (2020) |
|
| Camellia nitidissima var. microcarpa | 4.4 | 0.533 | – | – | – | Lu et al. (2019) | |
Similar to C. huana, many other yellow-flowered Camellia species, such as Camellia chrysanthoides H. T. Chang (Chen et al., 2019), Camellia micrantha S. Y. Liang et Y. C. Zhong (Chen et al., 2019), Camellia parvipetala J. Y. Liang et Su (Chen et al., 2019), Camellia nitidissima Chi (Li et al., 2019, Lu et al., 2020) and C. nitidissima Chi var. microcarpa Chang et Ye (Lu et al., 2019), exhibit a small distribution range and low level of genetic diversity (Table 5). Many natural populations have disappeared owing to excessive land use and transplantation. The number of individuals in the surviving natural populations has declined sharply (Chen et al., 2019, Li et al., 2019, Lu et al., 2019). The influence of human factors is not reflected in the results of the present study. In the future, the genetic diversity of these Camellia species may be further eroded by these adverse disturbances. It is therefore urgent to strengthen the protection of existing populations of these yellow-flowered Camellia species.
4.2. Genetic structure and differentiation of Camellia huanaGenetic differentiation between C. huana populations in this study was high (FST = 0.2159 for SSRs, FST = 0.9318 for cpDNA). The dispersal mechanism of a species can influence the level of gene flow and patterns of genetic structure among populations (Miao et al., 2017); in C. huana, this includes the migration and movement of both seeds and pollen. A previous study suggested that seeds of the congener C. japonica were mainly transmitted by gravity with limited movement (Ueno et al., 2000). Parentage analysis of seedlings of C. flavida in a 15 hm2 plot revealed a seed dispersal distance of 0.65–51.05 m (mean 12.47 m), with 81.0% of dispersed seeds found within 20 m (Peng and Tang, 2017). Our fieldwork found that C. huana seeds are also large and heavy, so we speculate that they disperse by gravity without long-distance movement, contributing to the limited cytoplasmic seed-mediated gene flow. Low gene flow (Nm = 0.073 for cpDNA) between populations also confirmed this inference. Although the pollen transmission mechanism of C. huana is still unclear, the related species Camellia petelotii (Merrill) Sealy receives frequent sunbird and honeybee visitors (Sun et al., 2017). C. flavida has a pollen dispersal distance of 3.06–194.73 m (mean 29.03 m), with 72.2% of dispersed pollen found within 20 m (Peng and Tang, 2017). The natural populations of C. huana grow in evergreen, broad-leaved forests on limestone. Limestone landforms are natural "terrestrial islands" exhibiting spatial isolation on restricted land masses (Gao et al., 2015). Therefore, the limestone habitat may have led to the discontinuous distribution of C. huana and prevented the transmission of seeds and pollen, resulting in restricted gene flow among populations and increased genetic differentiation. Although we identified two genetic groups consistent with geographical distribution using SSR markers, there was only one mutational step between the chloroplast haplotype (C3) of group I and the chloroplast haplotype (C5) of group II. Whether there is a morphological differentiation between members of these groups remains to be determined.
4.3. Conservation implicationsGenetic diversity is the driving force for adaptation to various changeable environments, and one of the main purposes of conservation genetics is to maintain the genetic diversity of threatened species (Lowe et al., 2005, Bhattacharyya and Kumaria, 2015). According to the results of our STRUCTURE analysis, the five natural C. huana populations should be split into two management units, and we should place more emphasis on in situ conservation. Since one management unit contains only the JSP population with about 200 individuals, we should give this population highest priority for conservation; the other management unit contains four populations (NJT, DLT, NRT and HLT). High levels of genetic differentiation among populations suggest that the transplanted populations originated from the neighboring limestone mountains, respectively. Therefore, some ex situ conservation measures are necessary for transplanted populations, with as many sample individuals as possible covering all populations.
Author contributionsShuang Li, Shang-Li Liu, Si-Yu Pei and Man–Man Ning collected plant samples. Shuang Li, Shang-Li Liu and Si-Yu Pei performed the experiments. Shuang Li analyzed the data and wrote the manuscript. Shao-Qing Tang designed and supervised the study, and also wrote and revised the manuscript. All authors read and approved the final manuscript.
Declaration of Competing InterestThe authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
AcknowledgementsThis study was supported by the National Natural Science Foundation of China (grant number 31760088) and Biodiversity Survey, Observation and Assessment Program of the Ministry of Ecology and Environment of China (grant number 837208).
Appendix A. Supplementary dataSupplementary data to this article can be found online at https://doi.org/10.1016/j.pld.2020.06.003.
Abbasov M., Akparov Z., Gross T., et al, 2018. Genetic relationship of diploid wheat(Triticum spp.) species assessed by SSR markers. Genet. Resour. Crop Evol, 65: 1441-1453. DOI:10.1007/s10722-018-0629-2 |
An M.T., 2005. Present status of the natural resource of camellias in Guizhou Province. Guizhou Forestry Sci. Tech, 33: 26-29. |
Bandelt H.J., Forster P., Röhl A., 1999. Median-joining networks for inferring intraspecific phylogenies. Mol. Biol. Evol, 16: 37-48. DOI:10.1093/oxfordjournals.molbev.a026036 |
Bhattacharyya P., Kumaria S., 2015. Molecular characterization of Dendrobium nobile Lindl., an endangered medicinal orchid, based on randomly amplified polymorphic DNA. Plant Systemat. Evol, 301: 201-210. DOI:10.1007/s00606-014-1065-1 |
Birky C.W., 2008. Uniparental inheritance of organelle genes. Curr. Biol, 18: R692-R695. DOI:10.1016/j.cub.2008.06.049 |
Chen H.L., Lu X.L., Ye Q.Q., et al, 2019. Genetic diversity and structure of three yellow Camellia species based on SSR markers. Guihaia, 39: 318-327. |
Chung M.Y., López-Pujol J., Son S., et al, 2018. Patterns of genetic diversity in rare and common orchids focusing on the Korean peninsula: implications for conservation. Bot. Rev, 84: 1-25. DOI:10.1007/s12229-017-9190-5 |
Coates D.J., Carstairs S., Hamley V.L., 2003. Evolutionary patterns and genetic structure in localized and widespread species in the Stylidium caricifolium complex (Stylidiaceae). Am. J. Bot, 90: 997-1008. DOI:10.3732/ajb.90.7.997 |
Cornuet J.M., Luikart G., 1996. Description and power analysis of two tests for detecting recent population bottlenecks from allele frequency data. Genetics, 144: 2001-2014. |
Doyle J.J., Doyle J.L., 1990. Isolation of plant DNA from fresh tissue. Focus, 12: 39-40. |
Edgar R.C., 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res, 32: 1792-1797. DOI:10.1093/nar/gkh340 |
Ellstrand N.C., Elam D.R., 1993. Population genetic consequences of small population size: implications for plant conservation. Annu. Rev. Ecol. Systemat, 2: 217-242. |
Evanno G., Regnaut S., Goudet J., 2005. Detecting the number of clusters of individuals using the software structure: a simulation study. Mol. Ecol, 14: 2611-2620. DOI:10.1111/j.1365-294X.2005.02553.x |
Excoffier L., Smouse P.E., Quattro J.M., 1992. Analysis of molecular variance inferred from metric distances among DNA haplotypes: application to human mitochondrial DNA restriction data. Genetics, 131: 479-491. |
Gao Y., Ai B., Kong H., et al, 2015. Geographical pattern of isolation and diversification in karst habitat islands: a case study in the Primulina eburnea complex. J. Biogeogr, 42: 2131-2144. DOI:10.1111/jbi.12576 |
Huang C.-C., Hung K.-H., Hwang C.-C., et al, 2011. Genetic population structure of the alpine species Rhododendron pseudochrysanthum sensu lato (Ericaceae)inferred from chloroplast and nuclear DNA. BMC Evol. Biol, 11: 108. DOI:10.1186/1471-2148-11-108 |
Levy E., Byrne M., Coates D.J., et al, 2016. Contrasting influences of geographic range and distribution of populations on patterns of genetic diversity in two sympatric Pilbara acacias. PloS One 11: e0163995. DOI:10.1371/journal.pone.0163995 |
Li X., Wang J., Fan Z., et al, 2019. Genetic diversity in the endangered Camellia nitidissima assessed using transcriptome-based SSR markers. Trees (Berl.), 34: 543-552. DOI:10.1007/s00468-019-01935-1 |
Librado P., Rozas J., 2009. DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics, 25: 1451-1452. DOI:10.1093/bioinformatics/btp187 |
Liu Y., Yang S., Ji P., et al, 2012. Phylogeography of Camellia taliensis (Theaceae)inferred from chloroplast and nuclear DNA: insights into evolutionary history and conservation. BMC Evol. Biol, 12: 92. DOI:10.1186/1471-2148-12-92 |
Liufu Y.-Q., Peng G.-Q., Lu Y.-B., et al, 2014. Development and characterization of 38 microsatellite markers for Camellia flavida based on transcriptome sequencing. Conserv. Genet. Resour, 6: 1007-1010. DOI:10.1007/s12686-014-0270-0 |
Logan S.A., Phuekvilai P., Sanderson R., et al, 2019. Reproductive and population genetic characteristics of leading-edge and central populations of two temperate forest tree species and implications for range expansion. For. Ecol.Manage, 433: 475-486. DOI:10.1016/j.foreco.2018.11.024 |
Lowe A.J., Boshier D., Ward M., et al, 2005. Genetic resource impacts of habitat loss and degradation; reconciling empirical evidence and predicted theory for neotropical trees. Heredity, 95: 255-273. DOI:10.1038/sj.hdy.6800725 |
Lu Y.-B., Liufu Y.-Q., Peng G.-Q., et al, 2014. Development of 21 microsatellite primers for Camellia pingguoensis (Theaceae) using 454 sequencing. Conserv.Genet. Resour, 6: 791-793. DOI:10.1007/s12686-014-0221-9 |
Lu X.L., Chen H.L., Liang X.Y., et al, 2019. Genetic diversity of peripheral population of Camellia nitidissima and variety microcarpa. Mol. Plant Breed, 17: 301-306. |
Lu X., Chen H., Wei S., et al, 2020. Chloroplast and nuclear DNA analyses provide insight into the phylogeography and conservation genetics of Camellia nitidissima (Theaceae) in southern Guangxi, China. Tree Genet. Genomes, 16: 8. DOI:10.1007/s11295-019-1390-1 |
Mantel N., 1967. The detection of disease clustering and a generalized regression approach. Cancer Res, 27: 209-220. |
Miao C.-Y., Yang J., Mao R.-L., et al, 2017. Phylogeography of Achyranthes bidentata(Amaranthaceae) in China's warm-temperate zone inferred from chloroplast and nuclear DNA: insights into population dynamics in response to climate change during the Pleistocene. Plant Mol. Biol. Rep, 35: 166-176. DOI:10.1007/s11105-016-1013-z |
Nybom H., 2004. Comparison of different nuclear DNA markers for estimating intraspecific genetic diversity in plants. Mol. Ecol, 13: 1143-1155. DOI:10.1111/j.1365-294X.2004.02141.x |
Peng G.Q., Tang S.Q., 2017. Fine-scale spatial genetic structure and gene flow of Camellia flavida, a shade-tolerant shrub in karst. Acta Ecol. Sin, 37: 7313-7323. |
Pons O., Petit R.J., 1996. Measuring and testing genetic differentiation with ordered versus unordered alleles. Genetics, 144: 1237-1245. |
Qin H.N., Yang Y., Dong S.Y., et al, 2017. Threatened species list of China's higher plants. Biodivers. Sci, 25: 696-744. DOI:10.17520/biods.2017144 |
Ramanatha Rao V., Hodgkin T., 2002. Genetic diversity and conservation and utilization of plant genetic resources. Plant Cell Tissue Organ Cult, 68: 1-19. DOI:10.1023/A:1013359015812 |
Rice W.R., 1989. Analyzing tables of statistical tests. Evolution, 43: 223-225. DOI:10.1111/j.1558-5646.1989.tb04220.x |
Rosenberg N.A., Burke T., Elo K., et al, 2001. Empirical evaluation of genetic clustering methods using multilocus genotypes from 20 chicken breeds. Genetics, 159: 699-713. |
Solórzano S., Arias S., Dávila P., 2016. Genetics and conservation of plant species of extremely narrow geographic range. Diversity, 8: 31. DOI:10.3390/d8040031 |
Spooner D.M., Núñez J., Trujillo G., et al, 2007. Extensive simple sequence repeat genotyping of potato landraces supports a major reevaluation of their gene pool structure and classification. Proc. Natl. Acad. Sci. U. S. A, 104: 19398-19403. DOI:10.1073/pnas.0709796104 |
Sun S.-G., Huang Z.-H., Chen Z.-B., et al, 2017. Nectar properties and the role of sunbirds as pollinators of the golden-flowered tea (Camellia petelotii). Am. J. Bot, 104: 468-476. DOI:10.3732/ajb.1600428 |
Ueno S., Tomaru N., Yoshimaru H., et al, 2000. Genetic structure of Camellia japonica L. in an old-growth evergreen forest, Tsushima, Japan. Mol. Ecol, 9: 647-656. DOI:10.1046/j.1365-294x.2000.00891.x |
Wei S.-J., Lu Y.-B., Ye Q.-Q., et al, 2017. Population genetic structure and phylogeography of Camellia flavida (Theaceae) based on chloroplast and nuclear DNA sequences. Front. Plant Sci, 8: 718. DOI:10.3389/fpls.2017.00718 |
Wright S., 1931. Evolution in Mendelian populations. Genetics, 16: 97-159. |
Xi Z., Ruhfel B.R., Schaefer H., et al, 2012. Phylogenomics and a posteriori data partitioning resolve the Cretaceous angiosperm radiation Malpighiales. Proc.Natl. Acad. Sci. U. S. A, 109: 17519-17524. DOI:10.1073/pnas.1205818109 |
Xie D.Z., Ya Z.G., Han J.Y., et al, 2014. Study of distribution and protection strategies of Camellia tianeensis. J. Green Sci. Technol: 89-91. |
Yang Y., Pan Y., Gong X., et al, 2010. Genetic variation in the endangered Rutaceae species Citrus hongheensis based on ISSR fingerprinting. Genet. Resour. Crop Evol, 57: 1239-1248. DOI:10.1007/s10722-010-9571-7 |
Yao M.-Z., Ma C.-L., Qiao T.-T., et al, 2012. Diversity distribution and population structure of tea germplasms in China revealed by EST-SSR markers. Tree Genet.Genomes, 8: 205-220. DOI:10.1007/s11295-011-0433-z |
Yoichi W., Tomaru N., 2014. Patterns of geographic distribution have a considerable influence on population genetic structure in one common and two rare species of Rhododendron (Ericaceae). Tree Genet. Genomes, 10: 827-837. DOI:10.1007/s11295-014-0723-3 |
Zaya D.N., Molano-Flores B., Feist M.A., et al, 2017. Assessing genetic diversity for the USA endemic carnivorous plant Pinguicula ionantha R. K. Godfrey (Lentibulariaceae). Conserv. Genet, 18: 171-180. DOI:10.1007/s10592-016-0891-9 |
Zhao D., Yang J., Yang S., et al, 2014. Genetic diversity and domestication origin of tea plant Camellia taliensis (Theaceae) as revealed by microsatellite markers. BMC Plant Biol, 12. |



