b. Key Laboratory for Plant Diversity and Biogeography of East Asia, Kunming Institute of Botany, Chinese Academy of Sciences (CAS), Kunming, 650201, China;
c. University of Chinese Academy of Sciences, 100049, Beijing, China;
d. Flower Research Institute, Yunnan Academy of Agriculture Sciences, Kunming, 650201, China
Gene flow between sympatrically occurring plants is very common when their reproductive isolation has not been completely formed (Abbott et al., 2013; Ma et al., 2010a, 2010b; Marczewski et al., 2015; Milne et al., 1999, 2003; Zha et al., 2008,2010). Gene flow is generally associated with successful pollen flow between species that have overlapping flowering times, mediated by wind in anemophilous plants, or by shared pollinators. Importantly, gene flow from a common species may accelerate the threat of extinction for rare species. For instance, pollen flow from other species can cause pollen and/or ovule discounting, and gene flow can lead to genetic swamping, by which partially fertile and viable hybrids replace threatened genotypes (Ma et al., 2019). Due to weak reproductive barriers among close relatives, hybridization has been frequently reported in sympatric distribution Rhododendron species (i.e. Milne and Abbott, 2008; Ma et al., 2010b; Zha et al., 2008; Yan et al., 2015). Indeed, natural hybridization is typically considered deleterious in plant conservation genetics, especially when rare species hybridize with widespread species of related taxa (e.g., Gilman and Behm, 2011; Burgess and Husband, 2006; Ma et al., 2016).
Rhododendron pubicostatum T. L. Ming is listed as an endangered species in the Red List of Rhododendrons (Gibbs et al., 2011). Only one population (Jiaozi Mountain) of Rh. pubicostatum was recorded from specimen records, and the total mature individuals of the species are less than 5000 based on our present investigations. It is consistent with the concept of Plant Species with Extremely Small Populations (PSESP), which focuses on species that face an elevated risk of extinction, characterized by small remaining populations in restricted habitats and exposure to serious human disturbances (Ma et al., 2013; Sun, 2016; Sun et al., 2019a, 2019b). During our fieldwork in 2017 , we found Rh. pubicostatum, Rh. bureavii Franchet and Rh. sikangense var. exquisitum T. L. Ming sympatric distribution in Jiaozi Mountain. The overlapping flowering periods of the three taxa and intermediate morphological characters of Rh. pubicostatum compared with Rh. bureavii and Rh. sikangense var. exquisitum suggested that Rh. pubicostatum might be hybrid origin. For instance, sparse hairs underside the leaf can be found in Rh. pubicostatum whereas nearly glandular in Rh. sikangense var. exquisitum and dense hairs in Rh. bureavii. Also Rh. pubicostatumhad numbers with 5-7 per inflorescence comparing with Rh. bureavii having 10- 20 flowers and Rh. sikangense var. exquisitum 3-6 flowers per inflorescence. In addition, there has been little research on the population structure and genetic diversity of Rh. pubicostatum, although this information is important for the development of meaningful conservation management strategies for this species.
In this study, we developed highly polymorphic microsatellites (SSRs) in Rh. pubicostatum and cross-amplified these SSRs in two sympatric Rhododendron species. By comparing genetic diversity and genetic compositions of the three taxa for clarifying the species status of Rh. pubicostatum. Our aim was to develop meaningful conservation strategies for Rh. pubicostatum.
2. Materials and methods 2.1. Plant materials and study siteRhododendron pubicostatum (subsect. Taliensia), was originally collected by P. I. Mao in 1952 from the Wumeng Mountains in Luquan County, Yunnan province at an altitude of 3650 m, but was not formally described as a new species until 1981 (Ming, 1981). There are no reports about Rh. pubicostatum until it was recalled as an endemic species in 2005 . Jiaozi mountain nature reserve (N 26.09°, E 102.84°) in the Wumeng Mountains is the only population of Rh. pubicostatum distributed, and the reserve is characterized by steep terrain, steep mountain and suspended altitude difference. There are abundant Rhododendron resources (e.g., Rh. pubicostatum, Rh. bureavii and Rh. sikangense var. exquisitum) in this area. Due the morphological traits of Rh. pubicostatum have the intermediate traits between Rh. bureavii and Rh. sikangense var. exquisitum. We suspect that Rh. pubicostatum is hybrid between Rh. sikangense var. exquisitum (subsect. Maculifera) and Rh. bureavii (subsect. Taliensia). In order to verify Rh. pubicostatum is a natural hybridization origin, we collected molecular experimental materials of Rh. pubicostatum, Rh. bureavii and Rh. sikangense var. exquisitum for the study.
In our study site, these two species are closely related both to each other and to Rh. pubicostatum. Although Rh. sikangense var. exquisitum in Jiaozi mountain nature reserve with the lowest elevation and Rh. bureavii in the highest, but they have much wider distribution than Rh. pubicostatum in the reserve. A total of 24 individuals randomly sampled from Rh. pubicostatum used to develop microsatellite markers were collected from the study site in Jiaozi mountain nature reserve, to assess polymorphisms of the developed microsatellite markers. The feasibility of the developed microsatellite markers was also assessed in 24 individuals from Rh. bureavii and Rh. sikangense var. exquisitum, respectively. Healthy young leaves were collected from randomly selected plants and immediately stored in silica-gel until DNA extraction. Voucher specimens were deposited in Yunnan Key Laboratory for Integrative Conservation of Plant Species with Extremely Small Populations, Kunming Institute of Botany under Chinese Academy of Sciences.
2.2. SSR primer development and amplification of DNA from the three sympatric Rhododendron speciesGenomic DNA was extracted from dried leaves by using the cetyltrimethylammonium bromide (CTAB) method as described by Doyle (1991). Extracted DNA from one sample each of the three species was sequenced in individual lanes on a MiSeq Benchtop sequencer (Illumina, Inc., San Diego, California, USA) using 2 × 250-bp read length. The expected products and primer sizes ranged from 100 to 280 bp and 18 to 23 bp, respectively. A summary of the microsatellite markers we developed are presented in Table 1.
| Locus name | Primer sequence (5'-3') | Repeat motif | Size (bp) | Ta (℃) |
| RH119455 | F:CCACTCTGGAAGACTCGTGTTR:ACTAGATAAATCATGTAGGTTTGGACC | (AC)8 | 201 | 59 |
| RH150193 | F:CCGGCCCACTACCTTCATAGR:GAACCAACCGGGATCCTACG | (AG)15 | 364 | 59 |
| RH16369 | F: AGGTGCAGCAAATTTCCGAA R: CCCTTCAATTCTCCTTTCTCAATCC | (AG)10 | 201 | 59 |
| RH175950 | F: TTCTCCATCGTTGCGAGACG R: CCTACCTCCTACACCGCAAG | (AG)8 | 201 | 59 |
| RH178203 | F: CAGCCAAACCCGAACCCTAA R: AAACCCAGAAGCCTCAGGTG | (aac)7 | 278 | 60 |
| RH185982 | F: GTCACTTCTATGAGTTCTATCGGT R: TCAAGATTCTGTGATTAATATCGCAT | (aat)7 | 201 | 59 |
| RH19511 | F: CCGGATAAGGTGAACGAGCT R: TCTGCAAATACAGTCTTCCCTTGA | (aac)9 | 201 | 60 |
| RH195227 | F: ATCATGAATCCCTTCCAACCC R:ACGTCTCCTTCTTGATCTTACTTCA | (AC)7 | 206 | 59 |
| RH19935 | F: ATACGCTATGTAGTGGGCCG R: GGAAACAGTGGCCGTTGGAT | (AG)10 | 201 | 61 |
| RH202706 | F: ACGAGGAAACGGAAGAATGGG R: GTTGCCAAGCCTCACACTTG | (AC)9 | 284 | 59 |
| RH209912 | F: TGCTCCTGACATGCCCTCTA R: GCCCATACCCTCAATAATTGGG | (aac)9 | 201 | 60 |
| RH226095 | F: ATTAATCTTGGTGGGCGCCA R: CCTCCTAGGCAAGAAACGTCA | (AG)9 | 346 | |
| Note: Ta=annealing temperature. | ||||
PCR amplifications were performed in a total reaction volume of 20 m L containing 25e50 ng of template DNA, 1 m L of each primer (0.6 mM), and 10 m L 2 × Taq PCR MasterMix [Tiangen (Tiangen Biotech, Beijing, China); 3 mM MgCl2, 100 mM KCl, 0.5 mM of each dNTP, 20 mM TriseHCl (pH 8.3), 0.1 U Taq polymerase]. PCR amplification was performed under the following conditions: an initial denaturation step for 3 min at 95 °C, followed by 33 cycles of denaturation at 94 °C for 30 s, annealing at 60 °C or 61 °C according to the Ta value (optimized for each locus; Table 1) for 30 s, extension at 72 °C for 45 s, and a final extension at 72 °C for 7 min. PCR amplification was carried out on a Veriti® 96-Well Thermal Cycler (Applied Biosystems, Foster City, USA). The PCR products were separated and visualized using an ABI3730 XL automated DNA fluorescent sequencer (Applied Biosystems, Foster City, CA, USA). Only products that were successfully amplified to an appropriate size and gave clear peaks were used for further polymorphism verification. From the 72 individuals studied, we validated a total of 12 microsatellite loci as polymorphic.
2.3. Comparative analysis of population genetics of three Rhododendron speciesWe visualized and quantified the SSR data using GENEMARKER v2.2.0 (Soft Genetics, State College, PA, USA). Based on the microsatellite data from all the sampled individuals, we then calculated certain population genetic diversity parameters, including number of alleles (NA), effective number of alleles (NE), Shannon information index (I), observed heterozygosity (HO), and expected heterozygosity (HE) using GenAlEx, version 6.502 (Peakall and Smouse, 2006). Deviation from HardyeWeinberg equilibrium (HWE) was calculated using the heterozygosity-based method (Nei's GIS) with 1000 permutations for each locus using the web version of GENEPOP, v4.0.10 (Raymond, 1995). Linkage disequilibrium (LD) for all loci pairs with probability adjusted using Bonferroni correction was also assessed using GENEPOP.
To further explore the genetic relationships among sampled individuals, principal coordinates analysis (PCoA) was performed in GenAlEx, version 6.502 (Peakall and Smouse, 2006). The pairwise genetic distances were based on Nei's standard genetic distance (Nei, 1978). Scatterplots were visualized in two-dimensional space for the first and second, as well as the first and third, principal coordinates.
To estimate the extent of admixture in the 72 study individuals, an analysis was conducted in STRUCTURE v.2.3.4 (Pritchard et al., 2000) using the Bayesian model-based clustering algorithm. Ten independent runs with a K value that varied from 1 to 10 were performed with a burn-in period of 1 × 104 steps followed by 1 × 105 Markov Chain Monte Carlo (MCMC) iterations, using an admixture model assuming correlated-allele frequencies. The bestfit number (K) of genetic clusters was estimated using STRUCTURE HARVESTER based on the maximum △K value (Earl, 2012; Evanno et al., 2005).
3. Results 3.1. Characteristics of SSRsA total of 66, 229 microsatellites were identified from sequences obtained from the three study individuals. Dinucleotides were the most abundant repeat motifs (54.7%, 36, 241 sequences), followed by tetranucleotides (22.3%, 14, 791 sequences), pentanucleotide (11.5%, 7614 sequences), hexanucleotide (5.9%, 3937 sequences) and trinucleotides (5.6%, 3646 sequences). The dominant dinucleotide motifs were GA/CT repeats (37.7%) and the least common repeat was GC/CG (0.3%). We designed 100 primers and used them to evaluate PCR amplification efficiency and polymorphs: 17 primers did not generate clear microsatellite peaks or failed to amplify DNA from any of the three species; 76, 71 and 64 primers successfully amplified DNA samples from Rh. pubicostatum, Rh. bureavii and Rh. sikangense var. exquisitum, respectively. Only 58 primers generated clear microsatellite peaks from samples of all three species. Therefore, 58 primers were used for further evaluation, of which 12 showed polymorphisms. Primer sequences are given in Table 1.
Of the 12 polymorphic SSR loci used in our study, four were dinucleotides, whereas the remainder were trinucleotides. The genetic diversity parameters of these three species are shown in Table 2. The numbers of alleles per locus ranged from 2 to 11 (mean 6.583). In Rh. pubicostatum, the observed and expected heterozygosities ranged from 0.125 to 0.958 (mean 0.517) and 0.119 to 0.821 (mean 0.565), respectively. Three polymorphic microsatellite loci (RH16369, RH175950 and RH226095) significantly deviated from HardyeWeinberg equilibrium in Rh. pubicostatum. In total, RH119455, RH150193, RH16369, RH175950, RH178203, RH195227, RH19935, RH202706, and RH226095 polymorphic microsatellite loci were detected to be deviated from HardyeWeinberg equilibrium (Table 2). Only three polymorphic microsatellite loci (RH185982, RH19511 and RH209912) did not deviate from HardyeWeinberg equilibrium in three study species, indicating the possibility of null alleles or an insufficient number of samples (Table 2). After Bonferroni correction, no significant linkage disequilibrium was observed for any pair of loci.
| Locus | Rh. pubicostatum | Rh. bureavii | Rh. sikangense var. exquisitum | |||||||||||||||
| NA | NE | I | HO | HE | PHWE | NA | NE | I | HO | HE | PHWE | NA | NE | I | HO | HE | PHWE | |
| RH119455 | 7 | 4.331 | 1.643 | 0.792 | 0.769 | 0.821 | 4 | 1.867 | 0.893 | 0.250 | 0.464 | 0.000*** | 5 | 3.176 | 1.425 | 0.708 | 0.731 | 0.003** |
| RH150193 | 10 | 5.592 | 1.973 | 0.833 | 0.821 | 0.947 | 12 | 8.170 | 2.242 | 0.667 | 0.878 | 0.003** | 3 | 1.976 | 0.815 | 0.500 | 0.494 | 0.990 |
| RH16369 | 7 | 3.986 | 1.645 | 0.583 | 0.749 | 0.050* | 5 | 2.824 | 1.234 | 0.292 | 0.646 | 0.000*** | 5 | 2.141 | 1.023 | 0.375 | 0.533 | 0.431 |
| RH175950 | 4 | 2.263 | 0.923 | 0.958 | 0.558 | 0.002** | 6 | 1.898 | 1.005 | 0.333 | 0.473 | 0.000*** | 8 | 2.141 | 1.220 | 0.417 | 0.533 | 0.000*** |
| RH178203 | 11 | 5.592 | 2.007 | 0.625 | 0.821 | 0.881 | 6 | 3.840 | 1.545 | 0.542 | 0.740 | 0.017* | 13 | 9.216 | 2.366 | 0.792 | 0.891 | 0.028* |
| RH185982 | 3 | 1.135 | 0.274 | 0.125 | 0.119 | 0.991 | 2 | 1.043 | 0.101 | 0.042 | 0.041 | 0.917 | 2 | 1.043 | 0.101 | 0.042 | 0.041 | 0.917 |
| RH19511 | 5 | 1.490 | 0.721 | 0.375 | 0.329 | 0.999 | 6 | 1.494 | 0.757 | 0.375 | 0.331 | 1.000 | 2 | 1.332 | 0.415 | 0.292 | 0.249 | 0.403 |
| RH195227 | 4 | 1.751 | 0.798 | 0.333 | 0.429 | 0.065 | 6 | 1.986 | 1.039 | 0.333 | 0.497 | 0.000*** | 6 | 1.759 | 0.867 | 0.375 | 0.431 | 0.000*** |
| RH19935 | 11 | 3.692 | 1.722 | 0.458 | 0.729 | 0.609 | 5 | 1.643 | 0.846 | 0.250 | 0.391 | 0.000*** | 12 | 6.160 | 2.105 | 0.375 | 0.838 | 0.000*** |
| RH202706 | 2 | 1.385 | 0.451 | 0.333 | 0.278 | 0.327 | 6 | 3.156 | 1.379 | 0.708 | 0.683 | 0.000*** | 5 | 1.629 | 0.806 | 0.292 | 0.386 | 0.066 |
| RH209912 | 7 | 1.572 | 0.854 | 0.417 | 0.364 | 1.000 | 4 | 1.297 | 0.514 | 0.250 | 0.229 | 0.998 | 8 | 3.740 | 1.641 | 0.917 | 0.733 | 0.738 |
| RH226095 | 8 | 5.512 | 1.834 | 0.375 | 0.819 | 0.000*** | 6 | 3.567 | 1.474 | 0.125 | 0.720 | 0.000*** | 7 | 3.681 | 1.531 | 0.542 | 0.728 | 0.562 |
| Note: Indices include allele number per locus (NA), number of effective alleles (NE), Shannon information index (I); observed heterozygosity (HO); expected heterozygosity (HE) and P value from exact tests for HardyeWeinberg equilibrium (PHWE) over 12 nuclear microsatellite loci. * Shows significant deviation from HardyeWeinberg equilibrium. *P < 0.05, **P < 0.01, ***P < 0.001. | ||||||||||||||||||
Population genetic analysis showed that Rh. pubicostatum had the slightly higher number of effective alleles (3.326) than the two other species. Rh. pubicostatum observed, expected, and unbiased expected heterozygosities were 0.486, 0.566 and 0.576, respectively. Pairwise Fst values of the three species were highest (0.231) between Rh. bureavii and Rh. sikangense var. exquisitum, and lowest (0.081) between Rh. pubicostatum and Rh. sikangense var. exquisitum (Table 3 and 4) (Fig. 1).
| Pop. | N | Nr | Np | NA | NE | I | HO | HE | UHe | F | LD |
| Rh. pubicostatum | 24 | 7.389 | 10 | 6.917 | 3.326 | 1.255 | 0.486 | 0.566 | 0.576 | 0.065 | 16 |
| Rh. bureavii | 24 | 7.997 | 14 | 6.617 | 2.753 | 1.124 | 0.350 | 0.518 | 0.526 | 0.262 | 14 |
| Rh. sikangense var. exquisitum | 24 | 7.000 | 15 | 7 | 3.325 | 1.231 | 0.472 | 0.557 | 0.566 | 0.113 | 26 |
| Note: number of individuals (N); allelic richness (Nr); number of private alleles (Np); number of alleles per locus (NA); number of effective alleles (NE); Shannon information index (I); observed heterozygosity (HO); expected heterozygosity (HE); unbiased expected heterozygosity=(2N/(2N-1)) * He (Uhe), fixation Index (F) and P value from exact tests for HardyeWeinberg equilibrium (PHWE) over 12 nuclear microsatellite loci. * Shows significant deviation from HardyeWeinberg equilibrium. *P < 0.05, **P < 0.01, ***P < 0.001. | |||||||||||
| Rh. pubicostatum | Rh. bureavii | Rh. sikangense var. exquisitum | |
| Rh. pubicostatum | 0.00000 | ||
| Rh. bureavii | 0.08323 | 0.00000 | |
| Rh. sikangense var. exquisitum | 0.08065 | 0.23123 | 0.00000 |
|
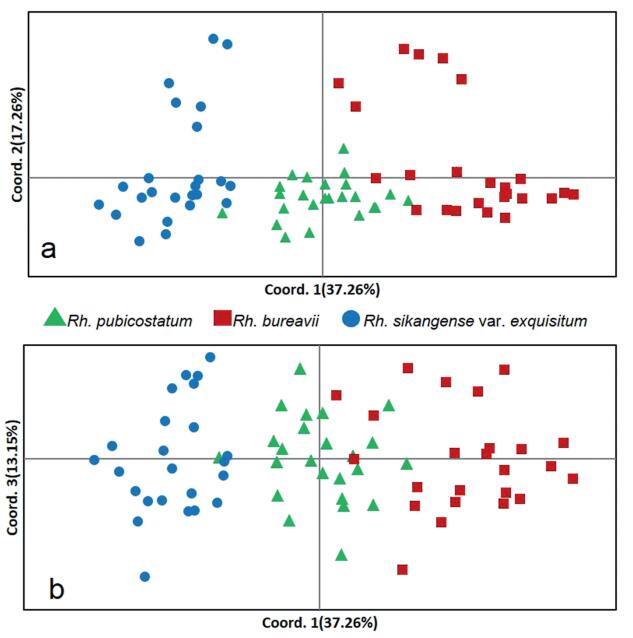
| Fig. 1 Principle coordinates analysis (PCoA) for 72 accessions of Rh. pubicostatum, Rh. bureavii and Rh. sikangense var. exquisitum based on genetic distances showing results for (a)the first and the second principle coordinates and (b) the first and the third principle coordinates. |
The two-dimensional PCoA plot showed that the first principal coordinate accounted for 37.26% of total variation and separates Rh. sikangense var. exquisitum from Rh. bureavii. Rh. pubicostatum samples are intermediate between those of Rh. bureavii and Rh. sikangense var. exquisitum (Fig. 2). The second and third principal coordinates (17.26% and 13.15% of total variation) did not provide sufficient information for distinguishing the three taxa.
|
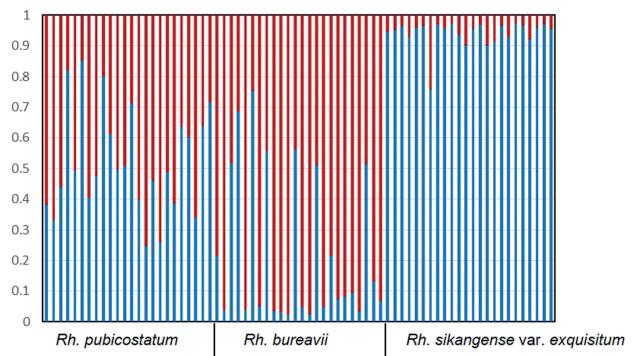
| Fig. 2 Results of STRUCTURE analysis for 72 samples based on 12 microsatellite loci. Samples are arranged according to morphological identification. The scale on the y axis represents the rate of the genetic composition for each sample in structure analysis. Red and blue represent the genetic composition of Rh. bureavii and Rh. sikangense var.exquisitum, respectively. |
Based on STRUCTURE analysis, the value of △K was clearly highest (13.336) when K=2. Following ten independent STRUCTURE runs with K=2, 23 individuals morphologically identified as Rh. sikangense var. exquisitum were assigned to one cluster with high probability (all q > 90%). The remaining single individual (no.1) seemed to be backcrossed Rh. sikangense var. exquisitum with some genetic materials from Rh. bureavii. Unlike Rh. sikangense var. exquisitum, more individuals (10/24) of Rh. bureavii showed evidence of admixture using the threshold value of q > 90%. These Rh. bureavii-like individuals were probably intermediate or backcrossed genotypes. The Rh. pubicostatum individuals have genetic material from both Rh. bureavii and Rh. sikangense var. exquisitum, with proportions of Rh. bureavii genetic material ranging from 40% to 60%.
4. Discussion 4.1. High efficiency of microsatellite development using MiSeqAlthough SSRs have been widely used in population genetics for numerous species, there are far fewer SSR polymorphic sites than in Miseq technology (Li et al., 2002; Glover et al., 2010; Guichoux et al., 2011). In the present study, multiple microsatellite loci were developed by using whole genome shotgun Illumina MiSeq sequencing, which is a useful tool for isolating new genetic markers in species with poor genomic resources and large genome sizes, while also circumventing the technical challenges of more traditional microsatellite enrichment protocols (Ekblom and Galindo, 2011; Ritchie et al., 2016). In high-throughput sequencing technology, RNA-seq technology is relatively lowcost and can result in large numbers of sites, but these tend to be non-neutral (Wang et al., 2009). Therefore, RNA-seq in not appropriate for obtaining the large numbers of neutral loci necessary for the genetic analysis of population structures. While RAD-seq technology and enzymatic digestion-based sequencing genotyping (GBS) can obtain more molecular marker sites than RNA-seq technology, the numbers of neutral sites revealed by these two technologies is still not comparable to that of Miseq technology (Mesak et al., 2014; Suchan et al., 2016). All 12 SSR primer pairs developed in this study gave an HE > 0.01 across all three Rhododendron taxa studied, suggesting that the sequences they amplify are polymorphic across these taxa (Ott, 1992), which is consistent with the finding that the three species have 100 percentage of polymorphic loci (PPL). Additionally, in Rh. pubicostatum the primers RH119455, RH150193, RH16369, RH178203, RH19935 and RH226095 resulted in amplification products showing an HE > 0.70 (Table 2), suggesting that these primers amplify highly polymorphic sequences (Ott, 1992). Miseq technology is indeed a relatively efficient and low-cost option for the development of a large number of neutral microsatellites.
4.2. Rhododendron pubicostatum maintains moderate levels of high genetic diversityMany factors affect the genetic diversity of species, including geographic distribution, breeding system, and population size (Hamrick and Godt, 1989). Species with larger ranges tend to have higher genetic diversity than those with limited ranges (Hamrick and Godt, 1989 Hamrick et al., 1992). The widespread species Rh. simsii Planch (HE=0.754), Rh. branchyacarpum G.Don (HE=0.815) and Rh. ripense Makino (HE=0.8) have higher genetic diversity than the narrowly endemic species Rh. pseudochrysanthum Hay. (HE=0.42) and Rh. oldhamii Maximowicz (HE=0.284) (Tan et al., 2009; Hirao, 2010; Kondo et al., 2009; Chen et al., 2014; Hsieh et al., 2013). In our study, Rh. sikangense var. exquisitum (HE=0.549) and Rh. bureavii (HE=0.508) showed lower levels of genetic diversity than were reported from the widespread species of Rh. simsii, Rh. branchyacarpum and Rh. ripense, which were analyzed with SSRs (Tan et al., 2009; Hirao, 2010; Kondo et al., 2009). Rh. bureavii and Rh. sikangense var. exquisitum showed quite low levels of genetic diversity for widespread species, but these figures are perhaps underestimates because we sampled only a single population of each taxon. Rh. pubicostatum (HE=0.565) showed higher genetic diversity than some narrowly endemic species have, including Rh. bureavii and Rh. oldhamii (Chen et al., 2014; Hsieh et al., 2013). Nybom (2004) calculated that the average diversity of endemic species when assessed using SSRs was 0.42, whereas that of widespread species was 0.62. From these numbers, the Rh. pubicostatum in our study would fall between endemic and widespread species, although the taxon is described as a narrowly endemic species known only from a single locality. But, in our study, Rh. pubicostatum showed higher levels of genetic diversity than Rh. bureavii and Rh. sikangense var. exquisitum. This is consistent with previous findings that the genetic diversity of natural hybrid individuals of 'Rh. ⅹ duclouxii' (HE=0.812) is higher than that of its parents Rh. spiciferum Franch. (HE=0.765) and Rh. spinuliferum Franch. (HE=0.728) (Yan et al., 2017). Hybridization or introgression between sympatric populations of species have been shown to increase genetic diversity (Dobes et al., 2004). In other words, these findings suggest that Rh. pubicostatum individuals are of a hybrid origin.
In the three sympatric taxa in our study, values of Ho are lower than HE, suggesting the occurrence of inbreeding depression. Although many plants appear to minimize self-pollination, resulting in deposition of more outcrossed pollen (Jong et al., 1993 ), it is possible that geitonogamy plays a major role in Rhododendron. In Rh. cyanocarpum and other Rhododendron species with known breeding systems (Ma et al., 2015; Li et al., 2018), geitogamous pollination is likely to occur frequently, due to large floral displays (comprising thousands of flowers) from a single individual and pollinator behavior, where the pollinators move between nearby flowers. Inbreeding depression might result from geitonogamy, a pollination strategy frequently reported in several Rhododendron (e.g., Rh. cynocarpun and Rh. longipedicellatum) (Ma et al., 2015; Liu et al., 2019). It has been proposed that geitonogamy is favored by the foraging behavior of pollinators, as most single Rhododendron individuals often display a large number of flowers and pollinators usually minimize inter-flower travel by preferentially foraging in adjacent flowers (Ma et al., 2015).
4.3. The hybrid origin of Rhododendron pubicostatum and implications for conservationBoth predominant components analysis (PCoA) and Bayes STRUCTURE analysis showed distinct genetic clusters formed by the Rh. bureavii and Rh. sikangense var. exquisitum plants sampled in our study, although gene flow clearly occurs between the two taxa. However, the Rh. pubicostatum individuals showed admixed ancestry, having characteristics of both Rh. bureavii and Rh. sikangense var. exquisitum, suggesting that they are hybrids of these two taxa. This provides further evidence of natural hybridization in Rhododendron, which has been frequently reported (e.g., Kron et al., 1993; Milne et al., 1999; Zhang et al., 2007; Zha et al., 2008; Ma et al., 2010b; Yan et al., 2015). Furthermore, the sympatric distribution, close relationship and overlapping flowering times of Rh. bureavii and Rh. sikangense var. exquisitum all provide opportunities for hybridization. Moreover, habitat disturbance has also been suggested to promote hybridization (Bleeker and Hurka, 2001; Hasselman et al., 2014). Our study area is a tourism hotspot, especially in April and May, when Rhododendrons flower. The construction of roads in this area may result in habitats where hybrid plants may outcompete parent taxa. However, unlike other known hybrid zones in Rhododendron where the parental species are dominant and only limited numbers of hybrids are observed (e.g., Rh. cyanocarpum < 20), Rh. pubicostatum is dominant in our study site, and the two parental species mainly occupy marginal areas. This hybrid zone is, therefore, unimodal, which implies that it may have been maintained over a long period, possibly before the development of Jiaozi Mountain for tourism.
If the Rh. pubicostatum individuals are of hybrid origin, this would also explain the higher genetic diversity observed in these specimens than that of the two parental species. Our preliminary conclusion that Rh. pubicostatum is of hybrid origin may not provide insight into whether Rh. pubicostatum requires protection. If strong evidence supports very early generations of Rh. pubicostatum and the existence offrequent introgressionwith parents, Rh.pubicostatumshouldnotbe a conservation priority. However, if evolutionary potential (e.g., differentiated local adaptation, formation of strong reproductive barriers between Rh. pubicostatum and its parents) can be detected in Rh. pubicostatum, it should be made a conservation priority (Oro et al., 2004; Genovart et al., 2007; Genovart, 2009). Therefore, addressing the conservation status of Rh. pubicostatum requires further study by employing more markers, samples as well as detailed examination of reproductive barriers among the three taxa.
Author's contributionsZhang X M performed the experiment and wrote the paper; Qin H T and Xie W J performed partly statistical analysis; Ma Y P and Sun W B designed the experiment and revised the paper.
Declaration of competing interestNone.
AcknowledgementsWe are grateful to Junbo Yang and Zhirong Zhang to providing technical assistance of lab work and data analysis. Laboratory work was performed at the Laboratory of Molecular Biology at the Germplasm Bank of Wild Species, Kunming Institute of Botany, Chinese Academy of Sciences. This work was supported by Science and Technology Basic Resources Investigation Program of China (Grant No. 2017FY100100), the Second Tibetan Plateau Scientific Expedition and Research (STEP) Program (Grant No. 2019QZKK0502), the Young Academic and Technical Leader Raising Foundation of Yunnan Province (No. 2018HB066), Yunnan Innovation Team Program for Conservation and Utilization of PSESP (Plant Species with Extremely Small Populations) (Grant No.2019HC015) and Applied Basic Research Project of Yunnan Province (Grant No. 2018BB010).
Abbott R., Albach D., Ansell S., et al, 2013. Hybridization and speciation. J. Evol.Biol, 26: 229-246. DOI:10.1111/j.1420-9101.2012.02599.x |
Bleeker W., Hurka H., 2001. Introgressive hybridization in Rorippa (Brassicaceae):gene flow and its consequences in natural and anthropogenic habitats. Mol.Ecol, 10(8): 2013-2022. DOI:10.1046/j.1365-294X.2001.01341.x |
Burgess K.S., Husband B.C., 2006. Habitat differentiation and the ecological costs of hybridization: the effects of introduced mulberry (Morus alba) on a native congener (M.rubra).. J. Ecol, 94: 1061-1069. DOI:10.1111/j.1365-2745.2006.01152.x |
Chen C.Y., Liang B.K., Chung J.D., et al, 2014. Demography of the upward-shifting temperate woody species of the Rhododendron pseudochrysanthum complex and ecologically relevant adaptive divergence in its trailing edge populations. Tree Genet. Genome, 10: 11-126. |
de Jong T.J., Waser N.M., Klinkhamer P.G., 1993. Geitonogamy: the neglected side of selfing. Trends Ecol. Evol, 8: 321-325. DOI:10.1016/0169-5347(93)90239-L |
Dobe#353; C.H. Mitchell-Olds T., Koch M.A., 2004. Extensive chloroplast haplotype variation indicates Pleistocene hybridization and radiation of North American Arabis drummondii, A. x divaricarpa, and A.holboellii (Brassicaceae).. Mol. Ecol, 13: 349-370. DOI:10.1046/j.1365-294X.2003.02064.x |
Doyle, J., 1991. DNA protocols for plantsdCTAB total DNA isolation. In: Hewitt, G.M., Johnston, A. (Eds.), Molecular Techniques in Taxonomy. Springer, Berlin, pp. 283-293.
|
Earl D.A., 2012. Structure HARVESTER: a website and program for visualizing STRUCTURE output and implementing the Evanno method. Conserv. Genet.Resour, 4: 359-361. DOI:10.1007/s12686-011-9548-7 |
Ekblom R., Galindo J., 2011. Applications of next generation sequencing in molecular ecology of non-model organisms. Heredity, 107: 1-15. DOI:10.1038/hdy.2010.152 |
Evanno G., Regnaut S., Goudet J., 2005. Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Mol. Ecol, 14: 2611-2620. DOI:10.1111/j.1365-294X.2005.02553.x |
Genovart M., 2009. Natural hybridization and conservation. Biodivers. Conserv, 18: 1435-1439. DOI:10.1007/s10531-008-9550-x |
Genovart M., Oro D., Juste J., Bertorelle G., 2007. What genetics tell us about the conservation of the critically endangered Balearic Shearwater? Biol. Conserv, 137: 283-293. DOI:10.1016/j.biocon.2007.02.016 |
Gibbs D., Chamberlain D., Argent G., 2011. The red list of Rhododendrons.. Botanic Gardens Conservation, 51. |
Gilman R.T., Behm J.E., 2011. Hybridization, species collapse, and species reemergence after disturbance to premating mechanisms of reproductive isolation. Evolution, 65: 2592-2605. DOI:10.1111/j.1558-5646.2011.01320.x |
Glover K.A., Hansen M.M., Lien S., et al, 2010. A comparison of SNP and STR loci for delineating population structure and performing individual genetic assignment. BMC Genet, 11: 2. |
Guichoux E., Lagache L., Wagner S., et al, 2011. Current trends in microsatellite genotyping. Mol. Ecol. Resour, 11: 591-611. DOI:10.1111/j.1755-0998.2011.03014.x |
Hamrick J., Godt M., BROWN M., et al, 1989. Plant Population Genetics, Breeding and Genetic Resources. Sinauer Associates, Sunderland, pp: pp. 43-63. |
Hamrick J., Godt M., Sherman-Broyles S., 1992. Factors influencing levels of genetic diversity in woody plant species. N. For, 6: 95-124. |
Hasselman D.J., Argo E.E., McBride M.C., et al, 2014. Human disturbance causes the formation of a hybrid swarm between two naturally sympatric fish species. Mol. Ecol, 23: 1137-1152. DOI:10.1111/mec.12674 |
Hirao A.S., 2010. Kinship between parents reduces offspring fitness in a natural population of Rhododendron brachycarpum. Ann. Bot, 105: 637-646. DOI:10.1093/aob/mcq018 |
Hsieh Y.C., Chung J.D., Wang C.N., et al, 2013. Historical connectivity, contemporary isolation and local adaptation in a widespread but discontinuously distributed species endemic to Taiwan, Rhododendron oldhamii (Ericaceae). Heredity, 111: 147-156. DOI:10.1038/hdy.2013.31 |
Kondo T., Nakagoshi N., Isagi Y., 2009. Shaping of genetic structure along Pleistocene and modern river systems in the hydrochoerus riparian azalea, Rhododendron ripense (Ericaceae). Am. J. Bot, 96: 1532-1543. DOI:10.3732/ajb.0800273 |
Kron K.A., Gawen L.M., Chase M.W., 1993. Evidence for introgression in azaleas(Rhododendron; Ericaceae): chloroplast DNA and morphological variation in a hybrid swarm on Stone Mountain, Georgia. Am. J. Bot, 80: 1095-1099. DOI:10.1002/j.1537-2197.1993.tb15335.x |
Li S.H., Sun W.B., Ma Y.P., 2018. Does the giant tree rhododendron need conservation priority? Glob. Ecol. Conserv, 15: e00421. |
Li Y.C., Korol A.B., Fahima T., et al, 2002. Microsatellites: genomic distribution, putative functions and mutational mechanisms: a review. Mol. Ecol, 11: 2453-2465. DOI:10.1046/j.1365-294X.2002.01643.x |
Liu D.T., Sun W.B., Ma Y.P., et al, 2019. Rediscovery and conservation of the critically endangered Rhododendron griersonianum in Yunnan, China.. Oryx, 53: 14. DOI:10.1017/S0030605318001278 |
Ma Y., Milne R.I., Zhang C., et al, 2010b. Unusual patterns of hybridization involving a narrow endemic Rhododendron species (Ericaceae) in Yunnan, China. Am. J. Bot, 97: 1749-1757. DOI:10.3732/ajb.1000018 |
Ma Y.P., Wu Z.K., Dong K., et al, 2015. Pollination biology of Rhododendron cyanocarpum (Ericaceae): an alpine species endemic to NW Yunnan, China. J. Systemat. Evol, 53: 63-71. DOI:10.1111/jse.12114 |
Ma Y.P., Xie W.J., Sun W.B., et al, 2016. Strong reproductive isolation despite occasional hybridization between a widely distributed and a narrow endemic Rhododendron species. Sci. Rep, 6. |
Ma Y., Chen G., Grumbine R.E., et al, 2013. Conserving Plant Species with Extremely Small Populations (PSESP) in China. Biodivers. Conserv, 22: 803-809. DOI:10.1007/s10531-013-0434-3 |
Ma Y., Marczewski T., Xue D., et al, 2019. Conservation implications of asymmetric introgression and reproductive barriers in a rare primrose species. BMC Plant Biol, 19. |
Ma Y.P., Zhang C.Q., Zhang J.L., et al, 2010a. Natural hybridization between Rhododendron delavayi and R.cyanocarpum (Ericaceae), from morphological, molecular and reproductive evidence.. J. Integr. Plant Biol, 52: 844-851. DOI:10.1111/j.1744-7909.2010.00970.x |
Marczewski T., Chamberlain D.F., Milne R.I., 2015. Hybridization in closely related Rhododendron species: half of all species-differentiating markers experience serious transmission ratio distortion. Ecol. Evol, 5: 3003-3022. DOI:10.1002/ece3.1570 |
Mesak F., Tatarenkov A., Earley R.L., et al, 2014. Hundreds of SNPs vs. dozens of SSRs: which dataset better characterizes natural clonal lineages in a selffertilizing fish? Front. Ecol. Evol, 2. |
Milne R.I., Abbott R.J., 2008. Reproductive isolation among two interfertile Rhododendron species: low frequency of post-F1 hybrid genotypes in alpine hybrid zones. Mol. Ecol, 17: 1108-1121. DOI:10.1111/j.1365-294X.2007.03643.x |
Milne R.I., Abbott R.J., Wolff K., et al, 1999. Hybridization among sympatric species of Rhododendron (Ericaceae) in Turkey: morphological and molecular evidence. Am. J. Bot, 86: 1776-1785. DOI:10.2307/2656674 |
Milne R.I., Terzioglu S., Abbott R.J., 2003. A hybrid zone dominated by fertile F1s:maintenance of species barriers in Rhododendron. Mol. Ecol, 12: 2719-2729. DOI:10.1046/j.1365-294X.2003.01942.x |
Ming T., 1981. New taxa of the genus Rhododendron from Yunnan-Sichuan. Acta Bot.Yunnanica, 3: 13-122. |
Nei M., 1978. Estimation of average heterozygosity and genetic distance from a small number of individuals. Genetics, 89: 583-590. |
Nybom H., 2004. Comparison of different nuclear DNA markers for estimating intraspecific genetic diversity in plants. Mol. Ecol, 13: 1143-1155. DOI:10.1111/j.1365-294X.2004.02141.x |
Oro D., Aguilar J.S., Igual Jos#233; Manuel, et al, 2004. Modelling demography and extinction risk in the endangered Balearic Shearwater. Biol. Conserv, 116: 93-102. DOI:10.1016/S0006-3207(03)00180-0 |
Ott J., 1992. Strategies for characterizing highly polymorphic markers in human gene mapping. Am. J. Hum. Genet, 51: 283. |
Peakall R.O.D., Smouse P.E., 2006. Genalex 6: genetic analysis in Excel. Population genetic software for teaching and research. Mol. Ecol. Notes, 6: 288-295. DOI:10.1111/j.1471-8286.2005.01155.x |
Pritchard J.K., Stephens M., Donnelly P., 2000. Inference of population structure using multilocus genotype data. Genetics, 155: 945-959. |
Raymond M., 1995. GENEPOP (version 1. 2): population genetics software for exact tests and ecumenicism. J. Hered, 86: 248-249. |
Ritchie H., Jamieson A.J., Piertney S.B., 2016. Isolation and characterization of microsatellite DNA markers in the deep-sea amphipod Paralicella tenuipes by Illumina MiSeq sequencing. J. Hered, 107: 367-371. DOI:10.1093/jhered/esw019 |
Suchan T., Pitteloud C., Gerasimova N.S., et al, 2016. Hybridization capture using RAD probes (hyRAD), a new tool for performing genomic analyses on collection specimens. PLos One, 11(3): e0151651. DOI:10.1371/journal.pone.0151651 |
Sun W.B., 2016. Words from the guest-in-chief. Plant Divers, 38: 207-208. DOI:10.1016/j.pld.2016.10.004 |
Sun W.B., Ma Y.P., Blackmore S., 2019a. How a new conservation action concept has accelerated plant conservation in China. Trends Plant Sci, 24: 4-6. DOI:10.1016/j.tplants.2018.10.009 |
Sun W.B., Yang J., Dao Z.L., 2019b. Study and Conservation of Plant Species with Extremely Small Populations (PSESP) in Yunnan Province, China. Science Press, Beijing, China. |
Tan X.X., Li Y., Ge X.J., 2009. Development and characterization of eight polymorphic microsatellites for Rhododendron simsii Planch (Ericaceae). Conserv.Genet, 10: 1553. DOI:10.1007/s10592-008-9791-y |
Wang Z., Gerstein M., Snyder M., 2009. RNA-Seq: a revolutionary tool for transcriptomics. Nat. Rev. Genet, 10: 57-63. DOI:10.1038/nrg2484 |
Yan L.J., Burgess K.S., Richard M., et al, 2017. Asymmetrical natural hybridization varies among hybrid swarms between two diploid Rhododendron species. Annals of Botany, 120: 51-61. DOI:10.1093/aob/mcx039 |
Yan L.J., Liu J., M#246;ller M., et al, 2015. DNA barcoding of Rhododendron (Ericaceae), the largest Chinese plant genus in biodiversity hotspots of the HimalayaeHengduan Mountain. Mol. Ecol. Resour, 15(4): 932-944. DOI:10.1111/1755-0998.12353 |
Zha H.G., Milne R.I., Sun H., 2008. Morphological and molecular evidence of natural hybridization between two distantly related Rhododendron species from the Sino-Himalaya. Bot. J. Linn. Soc, 156: 119-129. DOI:10.1111/j.1095-8339.2007.00752.x |
ZhaH.G., Milne R.I., Sun H., 2010. Asymmetric hybridization in Rhododendron agastum:a hybrid taxon comprising mainly F1s in Yunnan, China. Ann. Bot, 105: 89-100. DOI:10.1093/aob/mcp267 |
Zhang J.L., Zhang C.Q., Gao L.M., et al, 2007. Natural hybridization origin of Rhododendron agastum (Ericaceae) in Yunnan, China: inferred from morphological and molecular evidence. J. Plant Res, 120: 457-463. DOI:10.1007/s10265-007-0076-1 |



