b. CAS Key Laboratory for Plant Diversity and Biogeography of East Asia, Kunming Institute of Botany, Chinese Academy of Sciences, Kunming 650201, China;
c. University of Chinese Academy of Sciences, Beijing 100049, China
The Sino-Himalayan region is a temperate biodiversity hotspot with a high level of species endemism (Myers et al., 2000). Much of this diversity and endemism is attributed to the heterogeneous topography of the region and the diverse climates it supports (Shrestha et al., 2018). The formation of plant diversity in this region is synchronous with the uplift of the Qinghai-Tibet Plateau and has been intensified by climatic oscillations, especially the glaciale-interglacial cycle during the Quaternary (Muellner-Riehl et al., 2019; Sun et al., 2017; Wen et al., 2014; Wu, 1988). The mechanisms of plant speciation and diversification in this region are complicated and various (Muellner-Riehl et al., 2019; Wen et al., 2014). On the one hand, heterogeneous topography can create geographical barriers, which promote allopatric species divergence (e.g., Luo et al., 2017), and recent climatic oscillations can give rise to habitat fragmentation, which also interrupts the gene flow (e.g., Ma et al., 2017). On the other hand, divergent selection pressures across elevational gradients or heterogeneous habitats can also cause adaptive divergence in sympatry or parapatry (Funk et al., 2016), and even lead to adaptive radiation. This type of ecological divergence has been shown to be an essential speciation process for plants in the Sino-Himalayan region (e.g., Zhao et al., 2016).
Megacodon (Hemsl.) H. Smith is a small genus nested within a polytomy that contains Swertia L. and other genera in the Swertiinae (Chassot et al., 2001; Favre et al., 2016). However, Megacodon is morphologically distinct from other Swertiinae genera by having large campanulate flowers (Ho and Pringle, 1995). Megacodon contains two species: Megacodon stylophorus (C. B. Clark) H. Smith and Megacodon venosus (Hemsl.) H. Smith (Ho and Pringle, 1995). M. stylophorus is found at high elevations between 3000 and 4400 m a.s.l in western Sichuan, north-western Yunnan, southern Tibet of China, also in Bhutan, Nepal, and north-east India. M. venosus is endemic to China and is mainly found in Sichuan, western Hubei, and northern Chongqing at low to middle elevations between 600 and 2400 m a.s.l (Ho and Pringle, 1995). Previous studies of Megacodon have primarily focused on M. stylophorus, its phytochemistry (Liu et al., 2014), genetic diversity (Ge et al., 2005), embryology (Xue and Li, 2005), and breeding system (Meng et al., 2012). In contrast, M. venosus has been scarcely collected or reported. M. venosus was considered extinct until its rediscovery in 2007 by Prof. Zhen-Yu Li (Institute of Botany, the Chinese Academy of Sciences) in Northern Chongqing (Liu et al., 2014). Thus, it is vital to carry out investigations and conservation studies for this genus.
With rapid advances in next-generation sequencing (NGS), RNA-sequencing (RNA-seq) has become a more efficient approach for collecting gene sequences and studying ecological divergence in nonmodel organisms (Hudson, 2008; Strickler et al., 2012). For example, comparative transcriptome analysis can be used to identify candidate genes that underlie species diversification and adaptation, and then link these genes to specific biological processes (Voelckel et al., 2017). Previous studies have used genomeand transcriptome-based approaches to investigate adaptations to the high elevations of the Qinghai-Tibet Plateau region; however, few studies have examined these adaptations in plants, e.g., Crucihimalaya himalaica (Edgew.) Al-Shehbaz, O'Kane & R.A.Price (Qiao et al., 2016; Zhang et al., 2019), Primula poissonii Franchet and Primula wilsonii Dunn (Zhang et al., 2013), Cupressus gigantea W. C. Cheng & L. K. Fu and C. duclouxiana Silba (Ma et al., 2019), Salix brachista C. K. Schneider (Chen et al., 2019), Eutrema heterophyllum (W. W. Smith) H. Hara and E. yunnanense Franchet (Guo et al., 2018), Notopterygium incisum C. C. Ting ex H. T. Chang (Jia et al., 2019). For these studies, Gene Ontologies (GOs) of biotic and abiotic stress often provide novel insights into the local adaptation and species divergence in the extreme environments of the Qinghai-Tibet Plateau region.
During field investigation in 2017, we found a putative new species of Megacodon in Lushui county of north-west Yunnan at 1700 m a.s.l. This Lushui Megacodon has similar morphology and habitat as that of M. venosus; however, the Lushui Megacodon and M. venosus have extraordinarily disjunctive distributions. The new record of Megacodon from Lushui implies that the evolutionary history of this genus may be more complicated than previously thought. Two species co-occur in the Nushan Mountains, yet at different elevations. Furthermore, these observations suggest that the divergence of these Megacodon species may have been the result of both ecological divergence and allopatric speciation.
In this study, we described a new species of Megacodon from Lushui (Fig. 1) and determined its phylogenetic relationships within Megacodon. To identify genetic signatures of adaptive evolution to different elevations, we compared the transcriptome of M. stylophorus, which is distributed at high elevations, with that of the new species Megacodon lushuiensis, which is distributed at low elevations.
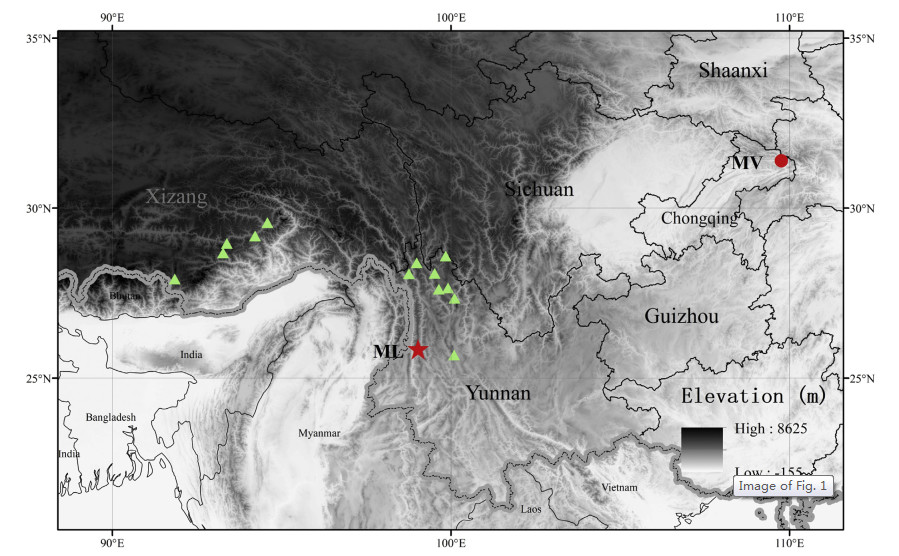
|
| Fig. 1 Geographic distributions of Megacodon samples. The red pentagram represents the putative new species (ML) in Lushui county, north-west Yunnan. The green triangles show the sampled natural populations of M. stylophorus. The red dot represents the M. venosus (MV) population from northern Chongqing. |
We sampled a total of 12 samples of M. stylophorus from different sites in Tibet and Yunnan, two samples of M. venosus from Chongqing, and two samples of the putative new species from Lushui, Yunnan (Fig. 1 and Table S1). Leaves were collected and dried in silica gel for DNA extraction. Specimens were deposited at the herbarium of Kunming Institute of Botany, Chinese Academy of Sciences (KUN). Flower organs of the Megacodon species from Lushui and M. stylophorus from Daxueshan Mountain were sampled by liquid nitrogen and stored at -80 ℃ for RNA-seq. Each sample was obtained from one individual.
2.2. DNA extraction, PCR amplification, and sequencingThe Plant Genomic DNA Kit (Tiangen Biotech, Beijing, China) was used to extract total genomic DNA following the manufacturer's protocol. The nuclear ribosomal internal transcribed spacer (nrITS) and two chloroplast markers, atpB-rbcL, and trnL-trnF, were amplified and sequenced following the protocols of previous studies (Favre et al., 2016; Matuszak et al., 2016). Primers used in this study for atpB-rbcL, trnL-trnF, and the nrITS region were the same as those used in previous studies (see Hoot et al., 1995; Taberlet et al., 1991; Grudinski et al., 2014 for the nrITS region). The 25 μL volume polymerase chain reactions (PCRs) contained 1.0 μL of genomic DNA (20 ng/μL), 0.3 μL of each primer (10 μM), 12.5 μL Ex Taq Version 2.0 (Takara, Dalian, China), and 10.9 μL of double-distilled H2O. PCRs were performed in a C1000 TouchTM Thermal Cycler (Bio-Rad, Hercules, CA, USA), with initiation at 95 ℃ for 2 min followed by 35 cycles of denaturation at 95 ℃ for 1 min, primer annealing at 53-56 ℃ for 1 min, primer extension at 72 ℃ for 1 min and a final extension step at 72 ℃ for 10 min. Alignments and subsequent manual adjustment were performed in BioEdit 7.2.5 (Hall, 1999) by using the multiple alignment software ClustalW (Thompson et al., 1994).
2.3. Phylogenetic analysesMaximum likelihood (ML) and Bayesian inference (BI) approaches were used for phylogenetic tree reconstruction with combined sequences from nrITS, atpB-rbcL, and trnL-trnF. Three Potalia Aubl. and three Lisianthius P. Browne species were set as outgroups (Table S2). In addition, 11 Gentiana L. species and 29 Swertia species were included as closely related genera (Table S2). ML analyses were performed on the CIPRES Science Gateway (www.phylo.org; Miller et al., 2010) through RAxML-HPC2 v.8.2.8 (Stamatakis, 2014) with 1000 bootstrap replicates. The best-fitting nucleotide substitution model for BI analyses was GTR + G, selected using the Akaike information criterion (AIC) in jModeltest 2.1.7 (Darriba et al., 2012). BI analyses were also performed on CIPRES using MrBayes v.3.2.6 (Ronquist et al., 2012) on XSEDE with the following settings: ngen = 10, 000, 000, samplefreq = 1000, printfreq = 1000 and sumt burnin = 2500.
2.4. Climatic data analysisOur fieldwork identified 15 precise locations where Megacodon is distributed. We used the 'raster' package to extract 19 bioclimatic variables for these sites from WorldClim (http://www.worldclim.org; Hijmans et al., 2005). Principal component analysis (PCA) of these environmental factors was performed by the dudi.pca function of the package 'ade4' in R 3.5.1 (R Core Team, 2018). We conducted paired two-tailed t-tests in Microsoft® Excel® 2016 (Microsoft Company, USA) to characterize factors that diverged between M. stylophorus and the other two Megacodon species (including the suspected new species from Lushui).
2.5. Transcriptome sequencing and functional annotationTRIzol reagent (Sigmae-Aldrich) was used to extract total RNA of samples following the manufacturer's instructions. A total amount of 3 μg of RNA per sample was used to construct a cDNA library with a fragment length range of 250-300 bp. The cDNA libraries were created with NEBNext® UltraTM RNA Library Prep Kit for Illumina® (NEB, USA), following the manufacturer's recommendations. Illumina Hiseq 2500 was used for sequencing the library preparations, and paired-end reads were generated. Library preparation and Illumina sequencing were performed at Novogene Bioinformatics Technology Co., Ltd., Beijing, China (www.novogene.cn). Clean data was obtained by trimming read adapter sequences, and filtering low-quality reads from raw data. De novo assembly of each individual was accomplished using the Trinity program (Raychowdhury et al., 2011) with min_kmer_cov set to 2, and all other parameters set to default.
Functions of all assembled unigenes were annotated based on the following databases: Nr (NCBI non-redundant protein sequences); Nt (NCBI non-redundant nucleotide sequences); Pfam (Protein family); KOG/COG (Clusters of Orthologous Groups of proteins); Swiss-Prot (A manually annotated and reviewed protein sequence database); KO (KEGG Ortholog database); and GO (Gene Ontology).
2.6. Homologous gene identified and positive selection analysisWe used OrthoFinder software v. 2.2.7 (Emms and Kelly, 2018, 2015) to identify homologous gene clusters (orthogroups). OrthoFinder was run with multiple sequences alignment (MSA) for gene tree inference and RAxML for tree inference method (-T ramxl -M msa). Orthogroups of single-copy genes (1:1 orthologs) were obtained for substitution rates estimation. Each orthogroup was aligned by Prank (Löytynoja and Goldman, 2005) at the condon level (with options "-F -codon"). To estimate a general evolutionary pattern of selective pressure for the two species, codeml program from PAML 4.9 (Yang, 2007) was used to calculate the dN, dS and dN/ dS with the free ratio model (model = 2, NSsites = 0) and ML pairwise alignment (runmode = -2). To reduce false-positive findings in subsequent evolutionary analyses, genes with length less than 150 bp, dS < 0.001 (to avoid a very large dN/dS ratio) and dS > 0.1 (to avoid suspicious paralogs) were discarded (Bustamante et al., 2005; Sun et al., 2018; Zhang et al., 2013).
Positive selection genes (dN/dS > 1) were used as the test set for the GO enrichment analysis, which was performed on web-based KOBAS software (Xie et al., 2011) by Fisher's exact test, while whole single-copy genes were selected as the background set. Significant enriched GO terms (P < 0.05) were selected and further analyzed using REVIGO (http://revigo.irb.hr/; Supek et al., 2011) to investigate in-depth GO similarities.
3. Results 3.1. Morphological comparisons and molecular variationComparative analysis of morphological traits indicates that the Megacodon species from Lushui is closely related to M. venosus (Fig. 2 and Table 1). The Megacodon species from Lushui is similar to M. venosus by its narrow leaf blades shape, calyx shape and the existence of glands at the base of calyx, but differs from M. venosus by its taller and thinner plant, lower stem leaf blades ellipticspathulate to elliptic-obovate (vs. elliptic-lanceolate), middle to upper stem leaf blades veins 3-5 (vs. 5-7), 11-15-flowered thyrses (vs. 7-11 flowered), and dense glands at the 1/3 base of the calyx (vs. sparse glands at the 1/2 base of the calyx). Two individuals from Lushui and two individuals from M. venosus differed by a total of eleven nucleotide substitutions and six insertions/deletions in the alignment dataset (including ITS, atpB-rbcL, trnL-trnF, Table S3).

|
| Fig. 2 Photographs of the putative new species Megacodon lushuiensis (A-E) and Megacodon venosus (F-J). A and F. Habitat and habit. B and G. Upper stem leaf blades. C and H. Middle stem leaves. D and I. Flower. E and J. Calyx. (A-D photographed by Jun-chu Peng; E-J photographed by Yi Yang). |
| M. lushuiensis | M. venosus | M. stylophorus | ||
| Habit | Plants 130-240 cm tall | Plants 45-85 (-180) cm tall | Plants 30-60 (-100) cm tall | |
| Stems | Basal to middle stems erect, terete, 1.1 | Stems erect, terete, stout, 1-1.5 cm in | Stems erect, terete, stout, 1-1.5 cm in | |
| -1.8 cm in diam at base; upper stems | diam | diam | ||
| slim 3-9 mm in diam | ||||
| Lower stem leaf blades | shape | elliptic-spathulate to elliptic-obovate | elliptic-lanceolate | elliptic to ovate-elliptic |
| veins | 5-7 | 5-7 | 7-9 | |
| Middle stem leaf blades | shape | elliptic-lanceolate to lanceolate | elliptic-lanceolate | elliptic to ovate-elliptic |
| size veins |
15-45 × 5-14 cm 3-5 |
10-30 × 3-6 cm 5-7 |
7-22 × 3-9 (-14) cm 7-9 |
|
| Upper stem leaf blades | shape | linear-lanceolate to linear | linear-lanceolate | ovate-lanceolate |
| size veins |
5-10 × 1.1-1.5 cm 3-5 |
5-7 × 0.7-1.2 cm 5-7 |
5e10 × 1.2-3 7-9 |
|
| Thyrses | 11-15-flowered | 7-11-flowered | 2-8-flowered | |
| Pedicel | 3-9 cm | 2.5-6 cm | 3-6 (-16) cm | |
| Calyx | lobes shape | elliptic-lanceolate, apex acuminate | elliptic-lanceolate, apex acuminate | oblong-spatulate, apex obtuse. |
| lobes length | 1.7-3.6 cm | 2.5-3 cm | 2-2.6 cm | |
| tube length | 4-6 mm | 2-3 mm | 6-8 mm | |
| Nectary | dense pustulose glands at the base ca. 1/ | Sparse pustulose glands at the base ca. | No glands | |
| 3 | 1/2 | |||
| Corolla | size lobes shape |
ca. 5-7 × 4-5 cm oblong-spatulate |
5-6 × 6-7.5 cm oblong-spatulate |
5-7 × 4-5 cm, oblong-spatulate |
| lobes length | 4-5.8 cm | 4-5 cm | 4-6 cm | |
| Anthers | 8-11 mm | 7-10 mm | 1-1.2 cm | |
| Style | ca. 1.7-2.0 cm | 1.5-1.8 cm | 1.5-1.8 cm | |
| Phenology | Fl. and fr. | Sep. to Nov. | Sep. to Nov. | Jun. to Sep. |
| Distribution | 1700 m, NW Yunnan | 600-2400 m. W Hubei, Sichuan, N | 3000-4400 m. S Sichuan, SE Tibet, NW | |
| Chongqing | Yunnan [Bhutan, NE India, Nepal] | |||
The data set of phylogenetic analyses comprised 62 accessions with 2383 characters. Because the topologies of phylogenetic trees from ML and BI analyses were mostly identical, only the ML tree is shown with the posterior probabilities (PP) from BI analysis (Fig. 3). Phylogenetic inference showed that Megacodon is most likely a monophyletic group (ML-BS = 100/BI-PP = 1.00 in Fig. 3). Twelve accessions of M. stylophorus from Tibet and Yunnan formed a wellsupported clade (96/1.00). In comparison, two accessions of M. venosus from Chongqing and two accessions from Lushui formed the other clade of Megacodon. The population of M. venosus and the species from Lushui each formed a monophyletic clade in this branch.
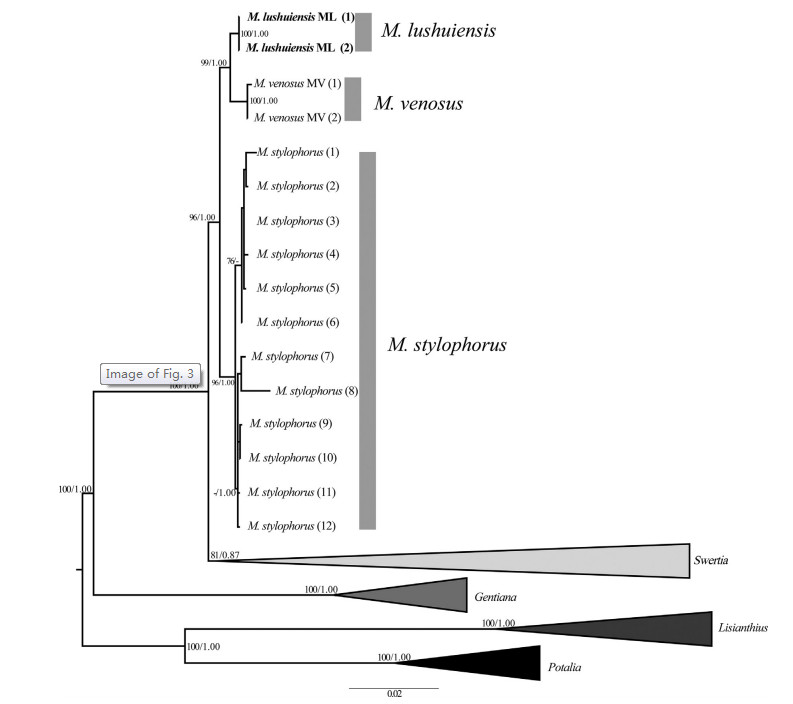
|
| Fig. 3 Molecular phylogeny of Megacodon and four other genera (Swertia, Gentiana, Lisianthius, and Potalia) based on nrITS, atpB-rbcL, and trnL-trnF sequences. ML bootstrap support values (BS) and Bayesian posterior probabilities (PP) are indicated on nodes and separated by a slash. Values for nodes with less than 75 (BS) or 0.85 (PP) are not shown. M. lushuiensis are shown bold. |
According to both morphological and molecular evidence, as well as the extraordinary disjunctive distribution with M. venosus, we propose the Megacodon plants from Lushui as a new species here.
M. lushuiensis J. C. Peng & H. Sun sp. nov.(Fig. 2A-E, and Fig. 4).
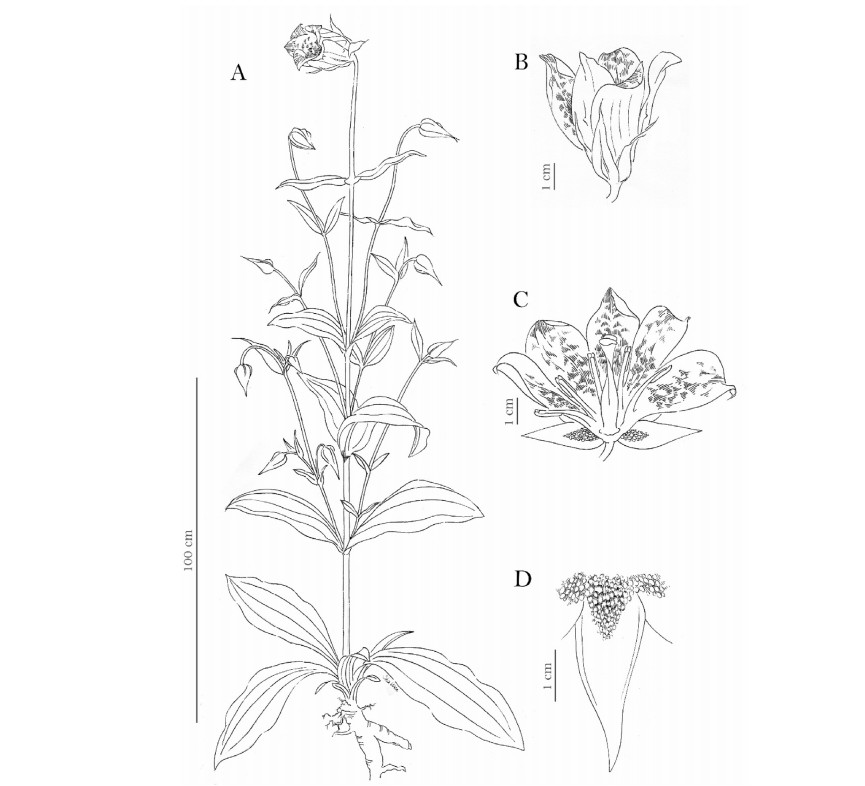
|
| Fig. 4 Megacodon lushuiensis J. C. Peng & H. Sun sp.nov. (drawn by Wen Sha). A. Habit B. Flower C. Opened corolla lobes D. Partial calyx. |
Holotype. China, Yunnan Province: Laowo Xiang, Lushui County, on the hillside, 1700 m a.s.l, 25.85°N, 99.03°E, flowering, 21 September 2017, J. C. Peng & L. Sun PM001 (KUN).
Paratype. The same locality, flowering and fruiting, 7 November 2017, J. C. Peng, L. Sun & Z. Chen PM002 (KUN).
Description. Plant perennial, herbaceous ca. 130-240 cm tall. Basal to middle stems erect, terete, 1.1-1.8 cm in diam at base; upper stems slim, 3-9 mm in diam. Lower stem leaves ellipticspathulate to elliptic-obovate, base cuneate and decurrent, veins 5-7 and arcuate; middle stem leaves elliptic-lanceolate to lanceolate, apex acuminate, 15-45 × 5-14 cm, veins 3-5 and arcuate; upper stem leaves linear-lanceolate to linear, 5-10 × 1.1-1.5 cm, veins 3-5 and arcuate. Thyrses 11-15-flowered. Pedicel 3-9 cm. Calyx campanulate, 1.7-3.6 cm, tube 4-6 mm; lobes ellipticlanceolate, apex acuminate, dense pustulous glands at the base ca. 1/3. Corolla white to pale yellow, with green veins, ca. 5-7 × 4-5 cm, tube 8-10 mm; lobes oblong-spatulate, 4-5.8 cm, apex rounded. Filaments flat and linear, 2.2-2.8 cm, alternate with lobes; anthers oblong-quadrate, 8-11 mm; Capsule ovoid-ellipsoid, 1.7-2.0 cm.
Phenology. Flowering and fruiting from Sep. to Nov.
Distribution and habitat. M. lushuiensis was found in Laowo Xiang of Lushui County, north-west Yunnan, south-west China (Fig. 1), growing on hillsides (north-west slope) and under shrub at ca. 1700 m a.s.l.
Conservation status. M. lushuiensis is only known from one population in Lushui county (criterion B2a), with an area of occupancy estimated to be less than 500 km2 (criterion B2b (i, ii)). The habitat of this population is near to cultivated field (less than 2 km) and suffering from invasive plants such as Ageratina adenophora (criterion B2b (iii)). The new species should be regarded as "Endangered" based on the IUCN (2012) criteria.
Etymology. The specific epithet refers to the type locality, Lushui County of Yunnan Province, in China.
3.4. Climatic nichePCA of 19 bioclimatic variables indicated clear climate niche differentiation between 13 M. stylophorus sites, one M. venosus site, and one M. lushuiensis site (Fig. 5). The first two PCs identified two climatic groups and explained 59% and 26.2% of the total variation, respectively (Fig. 5). The climatic niche of M. lushuiensis is closer to the population of M. venosus from Chongqing, whereas 13 sites of M. stylophorus formed another group. Among these 19 bioclimatic variables, 12 bioclimatic variables (BIO1, BIO5, BIO6, BIO8-BIO12, BIO14, BIO15, BIO17, and BIO19) showed significant differences between M. stylophorus and the other two species (P < 0.01; Table S4).

|
| Fig. 5 PCA analysis of bioclimatic factors of the putative new species (M. lushuiensis black square), M. stylophorus (red dots), and M. venosus (blue triangle). Each arrow represents a projection of a bioclimatic variable. |
We generated 55.2 and 55.3 million clean reads of RNA-seq data for M. lushuiensis and M. stylophorus, with a total size of 8.28 and 8.30 Gb, respectively (NCBI GenBank accession number: SRR11743766, SRR11743765 and Table 2). The median unigene length for M. lushuiensis after de novo assembly was 981 bp, with an N50 length of 2043 bp. The median unigene length for M. stylophorus was 1018 bp, with an N50 length of 2058 bp. A total of 24, 086 unigenes from M. lushuiensis and 23, 262 unigenes from M. stylophorus were annotated in at least one database with a significant match (e-value < 10-5).
| Species | Statistics of clean data | Frequency distribution of unigene lengths | ||||||||||||
| Clean Reads | Clean Bases (G) | Error (%) | Q20 (%) | Q30 (%) | GC Content (%) | Total number | min length (bp) | mean length (bp) | median length (bp) | max length (bp) | N50 (bp) | total nucleotides (bp) | ||
| M. lushuiensis | 55, 224, 736 | 8.28 | 0.03 | 96.3 | 90.2 | 43.2 | 35, 573 | 301 | 1400 | 981 | 16, 580 | 2043 | 49, 784, 866 | |
| M. stylophorus | 55, 301, 198 | 8.30 | 0.03 | 96.7 | 91.0 | 43.5 | 33, 202 | 301 | 1420 | 1018 | 15, 568 | 2058 | 47, 132, 153 | |
In this study, a total of 20, 679 and 20, 471 unigenes were detected in M. lushuiensis and M. stylophorus, respectively, and 23, 611 orthogroups were identified. Among these, 8926 orthogroups contained putative one-to-one single-copy genes between two species. To detect genes that are involved in ecological adaptation to high elevations, dN, dS, and dN/dS ratio of single-copy genes were calculated by PAML. A peak of dS distribution between M. stylophorus and M. lushuiensis was observed at 0.0411 ± 0.0210 (Fig. 6A). The dN/dS of most genes was less than one (Fig. 6B), which indicates purifying selection. Only 370 single-copy genes were identified as positively selected genes (PSGs, ω > 1) between two species.
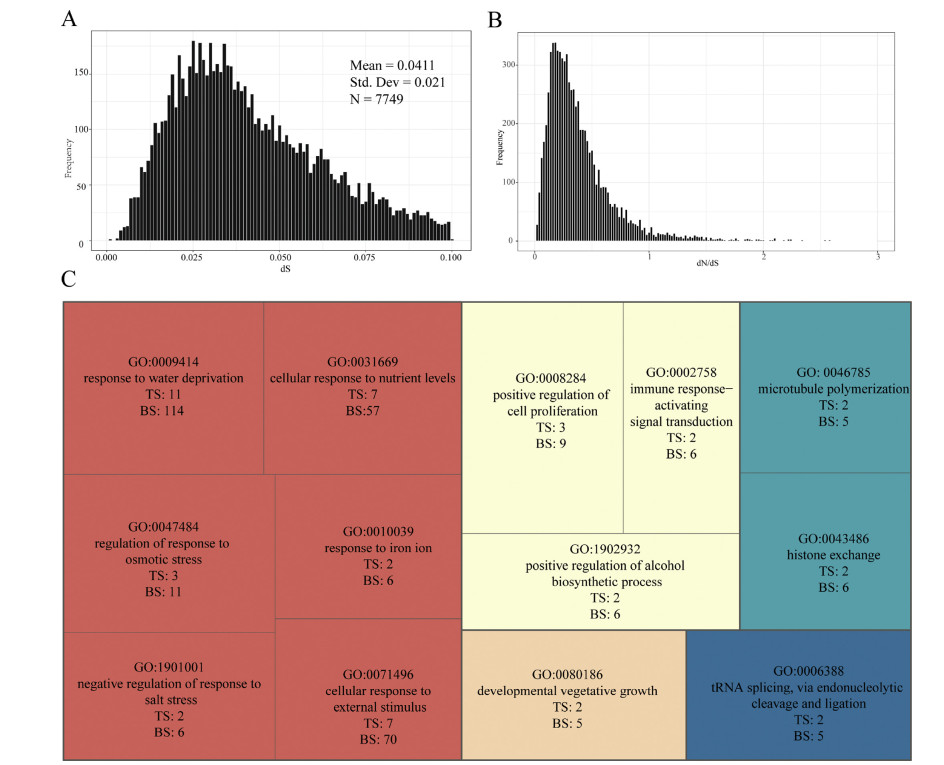
|
| Fig. 6 The dS (A) and dN/dS ratio (B) distribution of orthologs between M. stylophorus and M. lushuiensis. REVIGO treemap summarizing GO biological process categories overrepresented regarding ecological divergence of M. lushuiensis and M. stylophorus (C). KOBAS was used to identify GO biological processes that were overrepresented at p-values (Fisher's exact test)≤0.05. Similar colors indicate semantic similarity related to a similar functional category. The size of each rectangle is proportional to the p-value (Fisher's exact test) for that category. TS: frequency in the test set; BS: frequency in the background set. |
In enrichment analyses, the 370 positively selected genes were designated as the test sets, and 7749 single-copy genes (0.001 < dS < 0.1) were designated as the background set. Twentyfive GO categories were annotated to 49 pairs of positively selected genes, which were overrepresented (Fisher's exact test, p < 0.05) in the test sets (Table 3) by GO enrichment analysis in the KOBAS website. Then based on GO similarities, five clusters of biological processes were shown in the REVIGO treemap (Fig. 6C). A majority of biological processes in treemap were found to be related to specific adaptation traits, such as response to water (11 PSGs), regulation of response to osmatic (3 PSGs), cellular response to nutrient levels (7 PSGs), and cellular response to external stimulus (7 PSGs) (Fig. 6C, Table 3 and Table S5).
| Orthologs | Arabidopsis thaliana gene accession | Description | dN/dS |
| OG0007681 | AT1G32230 | RCD1, WWE proteineprotein interaction domain protein family | 1.99 |
| OG0010334 | AT3G17810 | PYD1, pyrimidine 1 | 1.31 |
| OG0002624 | AT2G39550 | PGGT-I, Prenyltransferase family protein | 1.05 |
| OG0009542 | AT1G11755 | LEW1, Undecaprenyl pyrophosphate synthetase family protein | 1.10 |
| OG0009326 | AT3G08040 | FRD3, MATE efflux family protein | 1.12 |
| OG0008777 | AT4G33630 | EX1, UvrB/UvrC domain protein | 1.15 |
| OG0009854 | AT3G63060 | EDL3, EID1-like 3 | 1.15 |
| OG0003758 | AT1G06770 | DRIP1, DREB2A-interacting protein 1 | 1.16 |
| OG0008137 | AT4G36020 | CSDP1, cold shock domain protein 1 | 1.15 |
| OG0009134 | AT5G28770 | BZO2H3, bZIP transcription factor family protein | 1.75 |
| OG0004371 | AT3G62420 | BZIP53, basic region/leucine zipper motif 53 | 1.42 |
| OG0006371 | AT3G48170 | ALDH10A9, aldehyde dehydrogenase 10A9 | 1.51 |
In this study, we described a new species of Megacodon, M. lushuiensis, and determined the phylogenetic relationships among Megacodon species. The three existing Megacodon species formed a well-supported monophyletic clade (Fig. 3). All samples of M. stylophorus formed a monophyletic clade. Furthermore, in our phylogenetic tree, M. lushuiensis is closely related to M. venosus, which is consistent with the morphological similarity between these two species. The phylogenetic relationship between Megacodon and four other lineages (Potalia, Lisianthius Gentiana, and Swertia) is congruent with previous studies (Favre et al., 2016). Although sharing many morphological similarities, M. lushuiensis can be easily distinguished from M. venosus by its leaf blade, inflorescence, extrafloral nectaries, and more. Importantly, the extrafloral nectaries in the basal portion of the calyx were neglected in previous morphological descriptions of M. venosus. The extrafloral nectaries of M. venosus and M. lushuiensis not only secrete nectar but also cover the petal and make it sticky (Fig. 2D, E, I, and J). We speculate that in addition to attracting ants to participate in ant-plant mutualisms, the secretions from M. venosus and M. lushuiensis extrafloral nectaries may directly participate in the protection of the flowers (Marazzi et al., 2013).
The exact distribution of M. lushuiensis in north-west Yunnan indicates that the most recent common ancestor of M. lushuiensis and M. venosus may occupy a more extensive area, from west Yunnan to central China. In the past, Megacodon populations from the Nujiang River valley may have been connected to M. venosus populations from central China through the eastern edge of the Qinghai-Tibet Plateau region or the Yunnan-Guizhou Plateau. The current fragmented distribution of this species pair may be the result of a vicariance event closely related to the climatic changes of the Sino-Himalayan region and its low reproductive capacity. Because of its humid climate and large elevational gradient, the Nujiang River valley harbors many distinct species that are identical to or sister to species in central or south China, such as Ichtyoselmis macrantha (Oliver) Lidén, Ypsilandra Franch., Asteropyrum peltatum (Franch.) Drumm. ex Hutch. The large elevational gradient and complex climate conditions in Nushan Mountain should provide more ecological opportunities for the maintenance of relict populations or species. Despite this, the fragmented distribution pattern of M. venosus and M. lushuiensis is still rare and may have important significance in biogeographical studies.
The occurrence of M. lushuiensis in north-west Yunnan also alters our understanding of the divergence of M. stylophorus and related species. Speciation of M. venosus and M. stylophorus was previously thought to have occurred through allopatric speciation, similar to that of Pleurospermum hookeri C. B. Clarke and P. giraldii Diels, which have a fragmented distribution between central China and the Qinghai-Tibet Plateau region (Bai et al., 2015). However, the fragmented distribution of M. lushuiensis and M. stylophorus occur at different elevations along the Nushan Mountain, suggesting that ecological differentiation may have played an important role in their divergence.
4.2. Functions under positive selection between M. lushuiensis and M. stylophorusTo further investigate why M. lushuiensis and M. stylophorus occupy distinct ecological niches along an elevational gradient, we used comparative transcriptome analysis. The majority of genes identified by GO enrichment of M. lushuiensis and M. stylophorus function in response to water deprivation (GO:0009414) and other external stimuli (e.g., GO:0047484 and GO:0071496) (Fig. 6C). These findings suggest that genes that respond to the environment may have been triggered by natural selection during ecological adaptation (Table 3). However, unlike many other alpine plants, especially alpine scree plants (Chen et al., 2019; Guo et al., 2018; Jia et al., 2019; Zhang et al., 2019), M. stylophorus does not appear to have adapted to high radiation. This may be because M. stylophorus grows in the forest or scrub habitat. Analysis of climatic variables revealed that M. stylophorus favors the colder and higher climatic drought conditions of high elevations (Fig. 5 and Table S1). In contrast, field observations indicate that M. lushuiensis prefers to grow in drier soil conditions such as semi-humid hillsides at low elevations (Fig. 2A). Habitat differentiation between these two species may have led some populations to experience water deficiency at different periods, which can influence plant growth and development. Our transcriptome analysis identified several genes (e.g., PGGT-1, ALDH10A9, and EDL3) under positive selection that is directly involved in water deprivation and hormone signaling pathways (Johnson et al., 2005; Koops et al., 2011; Missihoun et al., 2014). DRIP1 is a negative regulator in drought toleranceresponsive gene expression that was also found to be under positive selection (Qin et al., 2008). Our study also found that the following genes were under positive selection: LEW1, which response to endoplasmic reticulum stress, drought, and darkinduced senescence by catalyzing dolichol biosynthesis (Zhang et al., 2008); RCD1, which is involved in the regulation of stomatal conductance and the flavonoid accumulation pathway (Ahlfors et al., 2004; Suvorov et al., 2017); and CSDP1, which contains a cold shock domain and is involved in cold and salt stress responses (Yang and Karlson, 2013). The functions of these homologous genes emphasize that the genetic pathways that regulate efficient water use may have been important drivers of the divergence of M. lushuiensis and M. stylophorus.
Under a short growing season in the alpine region, M. stylophorus also require proper phenological responses. Our transcriptome analysis identified several genes that may play roles in these phenological responses. For example, in addition to its response to water deprivation, CSDP1 can also regulate seed germination (Yang and Karlson, 2013). Furthermore, bZIP53 is an enhancer for seed maturation gene transcription (Alonso et al., 2009), while BZOZH3 (also known as bZIP63) is involved in the regulation of a circadian oscillator gene PRR7 (Frank et al., 2018). The correct circadian clock guarantees that plants have photosynthetic advantages and can accurately anticipate environmental changes (Dodd et al., 2014, 2005). These homologous genes may strengthen the ability of M. stylophorus to respond to habitatspecific variability. Similarly, EX1 and FRD3, which were identified in our analysis, may help M. stylophorus tolerate higher oxidative stress and iron deficiency that result from radical-initiating factors at high elevations (Dogra et al., 2019; Lei et al., 2014), especially at the end of the growing period (Sakata et al., 2006). Overall, adapting to extreme environments (such as climatic drought, oxidative stress, and low temperature) in Sino-Himalayan regions may play an important role in the speciation of Megacodon.
5. ConclusionIn this study, we used field observations in conjunction with morphological and molecular data to propose a new Megacodon species from the Nujiang River valley, M. lushuiensis. The discovery of this species calls attention to the importance of ecological divergence between M. stylophorus and low-elevation Megacodon species. We also presented the phylogenetic position M. venosus for the first time and described a neglected trait in this species (sparse glands at 1/2 base of the calyx). Although population genetic and genomic studies are essential to understanding the demographic and divergence history of Megacodon, our analysis of the transcriptomes of M. stylophorus and M. lushuiensis provide valuable resources for the future genomic research and general insight into how ecological divergence occurred between these two Megacodon species. M. venosus and M. lushuiensis are both extremely rare in the field; we, therefore, recommend that conservation efforts be carried out timely and properly for these two species.
Author contributionsHS, XGM and YHW conceived and designed experiments. JCP performed field investigations, analyzed the data, and wrote the manuscript. XGM and HS revised the manuscript.
Declaration of Competing InterestWe declare that we do not have any commercial or associative interest that represents a conflict of interest in connection with the work submitted.
AcknowledgmentsThis study was supported by the Key Projects of the Joint Fund of the National Natural Science Foundation of China (U1802232), the Second Tibetan Plateau Scientific Expedition and Research (STEP) program (2019QZKK0502). Thanks to Si-Rong Yi (Chongqing Three Gorges Medical College) and Wei Zhang for providing valuable distribution information, Sha Wen for her line drawing of the new species, Ti-Cao Zhang (Yunnan University of Chinese Medicine) and Yan-Ting Hu for useful advice on RNA-seq analysis. We also appreciate the help of Jian Wen Zhang, Zhuo Zhou, Yang Niu, Yi Yang, Lu Sun, Zhe Chen, Bo Xu (Southwest Forestry University), Hong-Liang Chen, Li-Shen Qian, and Yong-Zeng Zhang during fieldwork.
Appendix A. Supplementary dataSupplementary data to this article can be found online at https://doi.org/10.1016/j.pld.2020.05.003.
Ahlfors R., Lång S., Overmyer K., et al, 2004. Arabidopsis RADICAL-INDUCED CELL DEATH1 belongs to the WWE proteineprotein interaction domain protein family and modulates abscisic acid, ethylene, and methyl jasmonate responses. Plant Cell, 16: 1925-1937. DOI:10.1105/tpc.021832 |
Alonso R., Oñate-Sánehez L., Weltmeier F., et al, 2009. A pivotal role of the basic leucine zipper transcription factor bZIP53 in the regulation of Arabidopsis seed maturation gene expression based on heterodimerization and protein complex formation. Plant Cell, 21: 1747-1761. DOI:10.1105/tpc.108.062968 |
Bai X., Ma X.G., Gao Y.D., et al, 2015. Intraspecific differentiation of Pleurospermum hookeri (Apiaceae), and its interspecific relationships with two close relatives in the genus Pleurospermum. J. Systemat. Evol, 53: 308-320. DOI:10.1111/jse.12144 |
Bustamante C.D., Fledel-Alon A., Williamson S., et al, 2005. Natural selection on protein-coding genes in the human genome. Nature, 437: 1153-1157. DOI:10.1038/nature04240 |
Chassot P., Nemomissa S., Yuan Y.M., et al, 2001. High paraphyly of Swertia L. (Gentianaceae) in the Gentianella-lineage as revealed by nuclear and chloroplast DNA sequence variation. Plant Systemat. Evol, 229: 1-21. |
Chen J.H., Huang Y., Brachi B., et al, 2019. Genome-wide analysis of cushion willow provides insights into alpine plant divergence in a biodiversity hotspot. Nat. Commun, 10: 1-12. DOI:10.1038/s41467-018-07882-8 |
Darriba D., Taboada G.L., Doallo R., et al, 2012. jModelTest 2: more models, new heuristics and high-performance computing Europe PMC Funders Group. Nat. Methods, 9: 772. |
Dodd A.N., Dalchau N., Gardner M.J., et al, 2014. The circadian clock has transient plasticity of period and is required for timing of nocturnal processes in Arabidopsis. New Phytol, 201: 168-179. DOI:10.1111/nph.12489 |
Dodd A.N., Salathia N., Hall A., et al, 2005. Cell biology: plant circadian clocks increase photosynthesis, growth, survival, and competitive advantage. Science, 309: 630-633. DOI:10.1126/science.1115581 |
Dogra V., Li Mingyue, Singh S., Li Mengping, Kim C., 2019. Oxidative posttranslational modification of EXECUTER1 is required for singlet oxygen sensing in plastids. Nat. Commun, 10: 1-12. |
Emms, D.M., Kelly, S., 2018. OrthoFinder2: fast and accurate phylogenomic orthology analysis from gene sequences. BioRxiv 466201.
|
Emms D.M., Kelly S., 2015. OrthoFinder : solving fundamental biases in whole genome comparisons dramatically improves orthogroup inference accuracy. Genome Biol, 16: 157. DOI:10.1186/s13059-015-0721-2 |
Favre A., Michalak I., Chen C.H., et al, 2016. Out-of-Tibet: the spatio-temporal evolution of Gentiana (Gentianaceae). J. Biogeogr, 43: 1967-1978. DOI:10.1111/jbi.12840 |
Frank A., Matiolli C.C., Viana A.J.C., et al, 2018. Circadian entrainment in Arabidopsis by the sugar-responsive transcription factor bZIP63. Curr. Biol, 28: 2597-2606. DOI:10.1016/j.cub.2018.05.092 |
Funk W.C., Murphy M.A., Hoke K.L., et al, 2016. Elevational speciation in action? Restricted gene flow associated with adaptive divergence across an altitudinal gradient. J. Evol. Biol, 29: 241-252. DOI:10.1111/jeb.12760 |
Ge X.J., Zhang L.B., Yuan Y.M., et al, 2005. Strong genetic differentiation of the east-himalayan Megacodon stylophorus (Gentianaceae) detected by inter-simple sequence repeats (ISSR). Biodivers. Conserv, 14: 849-861. DOI:10.1007/s10531-004-0655-6 |
Grudinski M., Pannell C.M., Chase M.W., et al, 2014. An evaluation of taxonomic concepts of the widespread plant genus Aglaia and its allies across Wallace's Line (tribe Aglaieae, Meliaceae). Mol. Phylogenet. Evol, 73: 65-76. DOI:10.1016/j.ympev.2014.01.025 |
Guo X., Hu Q., Hao G., et al, 2018. The genomes of two Eutrema species provide insight into plant adaptation to high altitudes. DNA Res, 25: 307-315. DOI:10.1093/dnares/dsy003 |
Hall, T.A., 1999. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. In: Nucleic Acids Symposium Series, pp. 95-98.
|
Hijmans R.J., Cameron S.E., Parra J.L., Jones P.G., Jarvis A., 2005. Very high resolution interpolated climate surfaces for global land areas. Int. J. Climatol, 25: 1965-1978. DOI:10.1002/joc.1276 |
Ho, T.N., Pringle, J.S., 1995. Gentianaceae. In: Wu, Z.-Y., Raven, P.H. (Eds.), Flora of China, vol. 16. Science Press and Missouri Botanical Garden, Beijing and St. Louis, pp. 1-139.
|
Hoot S.B., Culham A., Crane P.R., 1995. The utility of atpB gene sequences in resolving phylogenetic relationships: comparison with rbcL and 18S ribosomal DNA sequences in the Lardizabalaceae. Ann. Mo. Bot. Gard, 82: 194. DOI:10.2307/2399877 |
Hudson M.E., 2008. Sequencing breakthroughs for genomic ecology and evolutionary biology. Mol. Ecol. Resour, 8: 3-17. DOI:10.1111/j.1471-8286.2007.02019.x |
IUCN, 2012. In: IUCN Red List Categories and Criteria: Version 3.1, second ed. IUCN, Gland, Switzerland and Cambridge, UK. Iv + 32pp.
|
Jia Y., Bai J.Q., Liu M.L., et al, 2019. Transcriptome analysis of the endangered Notopterygium incisum: cold-tolerance gene discovery and identification of ESTSSR and SNP markers. Plant Divers, 41: 1-6. DOI:10.1016/j.pld.2019.01.001 |
Johnson C.D., Chary S.N., Chernoff E.A., et al, 2005. Protein geranylgeranyltransferase I is involved in specific aspects of abscisic acid and auxin signaling in Arabidopsis. Plant Physiol, 139: 722-733. DOI:10.1104/pp.105.065045 |
Koops P., Pelser S., Ignatz M., et al, 2011. EDL3 is an F-box protein involved in the regulation of abscisic acid signalling in Arabidopsis thaliana. J. Exp. Bot, 62: 5547-5560. DOI:10.1093/jxb/err236 |
Lei G.J., Zhu X.F., Wang Z.W., et al, 2014. Abscisic acid alleviates iron deficiency by promoting root iron reutilization and transport from root to shoot in Arabidopsis. Plant Cell Environ, 37: 852-863. DOI:10.1111/pce.12203 |
Liu C., Liao Z.X., Liu S.J., et al, 2014. Two new 2, 3-seco-hopane triterpene derivatives from Megacodon stylophorus and their antiproliferative and antimicrobial activities. Planta Med, 80: 936-941. DOI:10.1055/s-0034-1368612 |
Löytynoja A., Goldman N., 2005. An algorithm for progressive multiple alignment of sequences with insertions. Proc. Natl. Acad. Sci, 102: 10557-10562. DOI:10.1073/pnas.0409137102 |
Luo D., Xu B., Li Z.M., et al, 2017. The 'Ward LineeMekongeSalween Divide' is an important floristic boundary between the eastern Himalaya and Hengduan mountains: evidence from the phylogeographical structure of subnival herbs Marmoritis complanatum (Lamiaceae). Bot. J. Linn. Soc, 185: 482-496. DOI:10.1093/botlinnean/box067 |
Ma X.G., Sun W.G., Zhu W.D., et al, 2017. Resolving the phylogenetic relationships and evolutionary history of the East Asian endemic genus Rodgersia (Saxifragaceae) using multilocus data. Perspect. Plant Ecol, 25: 20-28. DOI:10.1016/j.ppees.2016.12.005 |
Ma Y., Wang J., Hu Q., et al, 2019. Ancient introgression drives adaptation to cooler and drier mountain habitats in a cypress species complex. Commun. Biol, 2: 213. DOI:10.1038/s42003-019-0445-z |
Marazzi B., Bronstein J.L., Koptur S., 2013. The diversity, ecology and evolution of extrafloral nectaries: current perspectives and future challenges. Ann. Bot, 111: 1243-1250. DOI:10.1093/aob/mct109 |
Matuszak S., Muellner-Riehl A.N., Sun H., et al, 2016. Dispersal routes between biodiversity hotspots in Asia: the case of the mountain genus Tripterospermum (Gentianinae, Gentianaceae) and its close relatives. J. Biogeogr, 43: 580-590. DOI:10.1111/jbi.12617 |
Meng L.H., Wang Y., Luo J., et al, 2012. Pollination ecology and its implication for conservation of an endangered perennial herb native to the East-Himalaya, Megacodon stylophorus (Gentianaceae). Plant Ecol. Evol, 145: 356-362. DOI:10.5091/plecevo.2012.684 |
Miller, M.A., Pfeiffer, W., Schwartz, T., 2010. Creating the CIPRES Science Gateway for inference of large phylogenetic trees. In: 2010 Gateway Computing Environments Workshop. GCE), pp. 1-8.
|
Missihoun T.D., Willèe E., Guegan J.P., et al, 2014. Overexpression of ALDH10A8 and ALDH10A9 genes provides insight into their role in glycine betaine synthesis and affects primary metabolism in Arabidopsis thaliana. Plant Cell Physiol, 56: 1798-1807. |
Muellner-Riehl A.N., Schnitzler J., Kissling W.D., et al, 2019. Origins of global mountain plant biodiversity: testing the 'mountain-geobiodiversity hypothesis'. J. Biogeogr, 46: 2826-2838. DOI:10.1111/jbi.13715 |
Myers N., Mittermeier R.A., Mittermeier C.G., et al, 2000. Biodiversity hotspots for conservation priorities. Nature, 403: 853-858. DOI:10.1038/35002501 |
Qiao Q., Wang Q., Han X., et al, 2016. Transcriptome sequencing of Crucihimalaya himalaica (Brassicaceae) reveals how Arabidopsis close relative adapt to the Qinghai-Tibet Plateau. Sci. Rep, 6: 1-8. |
Qin F., Sakuma Y., Tran L.S.P., et al, 2008. Arabidopsis DREB2A-interacting proteins function as Ring E3 ligases and negatively regulate plant drought stressresponsive gene expression. Plant Cell, 20: 1693-1707. DOI:10.1105/tpc.107.057380 |
R Core Team, 2018. R: A Language and Environment for Statistical Computing.
|
Raychowdhury R., Gnirke A., Fan L., et al, 2011. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol, 29: 644-652. DOI:10.1038/nbt.1883 |
Ronquist F., Teslenko M., Van Der Mark P., et al, 2012. MrBayes 3. 2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol, 61: 539-542. DOI:10.1093/sysbio/sys029 |
Sakata T., Nakano T., Yokoi Y., 2006. Altitudinal changes in Rubisco and APX activities in Aconogonum weyrichii in the alpine region of Mt. Fuji. Polar Biosci: 115-122. |
Shrestha N., Su X., Xu X., et al, 2018. The drivers of high Rhododendron diversity in south-west China: does seasonality matter?. J. Biogeogr, 45: 438-447. DOI:10.1111/jbi.13136 |
Stamatakis A., 2014. RAxML version 8: a tool for phylogenetic analysis and postanalysis of large phylogenies. Bioinformatics, 30: 1312-1313. DOI:10.1093/bioinformatics/btu033 |
Strickler S.R., Bombarely A., Mueller L.A., 2012. Designing a transcriptome nextgeneration sequencing project for a nonmodel plant species. Am. J. Bot, 99: 257-266. DOI:10.3732/ajb.1100292 |
Sun H., Zhang J.W., Deng T., et al, 2017. Origins and evolution of plant diversity in the Hengduan Mountains, China. Plant Divers, 39: 161-166. DOI:10.1016/j.pld.2017.09.004 |
Sun Y.B., Fu T.T., Jin J.Q., et al, 2018. Species groups distributed across elevational gradients reveal convergent and continuous genetic adaptation to high elevations. Proc. Natl. Acad. Sci, 115: E10634-E10641. DOI:10.1073/pnas.1813593115 |
Supek, F., Bošnjak, M., Škunca, N., et al., 2011. Revigo summarizes and visualizes long lists of gene ontology terms. PloS One 6.
|
Suvorov A., Jensen N.O., Sharkey C.R., et al, 2017. Opsins have evolved under the permanent heterozygote model: insights from phylotranscriptomics of Odonata. Mol. Ecol, 26: 1306-1322. DOI:10.1111/mec.13884 |
Taberlet P., Gielly L., Pautou G., et al, 1991. Universal primers for amplification of three non-coding regions of chloroplast DNA. Plant Mol. Biol, 17: 1105-1109. DOI:10.1007/BF00037152 |
Thompson J.D., Higgins D.G., Gibson T.J., 1994. Clustal W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res, 22: 4673-4680. DOI:10.1093/nar/22.22.4673 |
Voelckel C., Gruenheit N., Lockhart P., 2017. Evolutionary transcriptomics and proteomics: insight into plant adaptation. Trends Plant Sci, 22: 462-471. |
Wen J., Zhang J.Q., Nie Z.L., et al, 2014. Evolutionary diversifications of plants on the Qinghai-Tibetan plateau. Front. Genet, 5: 1-16. |
Wu Z.Y., 1988. Hengduan mountain flora and her significance. J. Jpn. Bot, 63: 297-311. |
Xie C., Mao X., Huang J., et al, 2011. KOBAS 2. 0: a web server for annotation and identification of enriched pathways and diseases. Nucleic Acids Res, 39: W316-W322. DOI:10.1093/nar/gkr483 |
Xue C.Y., Li D.Z., 2005. Embryology of Megacodon stylophorus and Veratrilla baillonii (Gentianaceae): descriptions and systematic implications. Bot. J. Linn. Soc, 147: 317-331. DOI:10.1111/j.1095-8339.2005.00371.x |
Yang Y., Karlson D., 2013. AtCSP1 regulates germination timing promoted by low temperature. FEBS Lett, 587: 2186-2192. DOI:10.1016/j.febslet.2013.05.039 |
Yang Z., 2007. PAML 4: phylogenetic analysis by maximum likelihood. Mol. Biol. Evol, 24: 1586-1591. DOI:10.1093/molbev/msm088 |
Zhang H., Ohyama K., Boudet J., et al, 2008. Dolichol biosynthesis and its effects on the unfolded protein response and abiotic stress resistance in Arabidopsis. Plant Cell, 20: 1879-1898. DOI:10.1105/tpc.108.061150 |
Zhang L., Yan H.F., Wu W., et al, 2013. Comparative transcriptome analysis and marker development of two closely related Primrose species (Primula poissonii and Primula wilsonii). BMC Genom, 14: 329. DOI:10.1186/1471-2164-14-329 |
Zhang T., Qiao Q., Novikova P.Y., et al, 2019. Genome of Crucihimalaya himalaica, a close relative of Arabidopsis, shows ecological adaptation to high altitude. Proc. Natl. Acad. Sci, 116: 7137-7146. DOI:10.1073/pnas.1817580116 |
Zhao J.L., Gugger P.F., Xia Y.M., et al, 2016. Ecological divergence of two closely related Roscoea species associated with late Quaternary climate change. J. Biogeogr, 43: 1990-2001. DOI:10.1111/jbi.12809 |



