b. School of Life Science, Engineering Research Center of Sustainable Development and Utilization of Biomass Energy, Yunnan Normal University, Kunming 650500, China;
c. University of the Chinese Academy of Sciences, Beijing 100049, China
Plant karyotype research is important for understanding the origin and evolution plant species, molecular phylogeny, and floristic geography (Sun et al., 2019). Karyomorphological study provides insight into potential evolutionary characteristics of kar-yotypes, as well as the cytological mechanisms driving the evolu-tion of plant diversity on a phylogenetic scale. In addition, karyomorphological study is a fast, inexpensive approach to classify plant species by identifying the basic cytological parameters of a species, including chromosome number, ploidy level, karyotype asymmetry, and karyotype coefficient of variation (Guerra, 2008). The number of chromosomes and karyotype of a species are stable characteristics that can reflect its basic genetic information.Furthermore, differences in chromosome numbers between pop-ulations is important evidence for determining reproductive isolation. Investigating the cytological features of species (e.g., via chromosome atlas or polyploid frequency) at a regional or flora scale may uncover the influence of geo-ecological environmental shifts on chromosome ploidy.
Various hypotheses have been proposed to explain the origin and distribution of plant polyploids. The prevailing hypothesis is that the doubled genome of polyploids renders them more adapt-able than diploids to extreme environments such as those encountered at high latitudes and high altitudes (Soltis and Soltis, 2000). For example, the region with the highest proportion of plant polyploids in the word is the Arctic Circle. Analysis of plant chromosome numbers in the Arctic indicates that the chromosome polyploid ratio in this region is more than 80% (Löve and Löve 1975). In addition, previous research has shown that the ratio of polyploid species at high altitude environments is relatively high (Löve and Löve 1967). Similarly, research has indicated that the polyploid frequency is much higher in the mountains than in the lowlands (Hanelt, 1966). The Qinghai-Tibet Plateau region is a well-known biodiversity hotspot that harbors numerous endemic plant species in extreme alpine environments. Although understanding species diversity in this area has been a long-term concern of sci-entists (Spicer, 2017; Sun et al., 2017), the underlying forces that have driven such species richness remains unclear. One potential explanation for the high level of endemic plant diversity in the Qinghai-Tibet Plateau is that increased polyploidization. Con-structing regional chromosome data is a promising approach to shedding light on the formation and evolutionary history of this flora.
The Brassicaceae family comprises c. 4000 species including economically important crops and the model plant Arabidopsis thaliana (L.) Heynh. (Appel and Al-Shehbaz, 2003; Kiefer et al., 2014). Brassicaceae contains approximately 102 genera and 412 species in China (Zhou et al., 2001). Anzhengxia Al-Shehbaz & D. A. German, Shangrilaia Al-Shehbaz, J. P. Yue & H. Sun, Baimashania Al-Shehbaz are three recently established endemic genera of Brassi-caceae from Qinghai-Tibet Plateau (Fig. 1). Anzhengxia (A. yechengnica (C. H. An) Al-Shehbaz & D. A. German) and Shangrilaia (Shangrilaia nana Al-Shehbaz, J. P. Yue & H. Sun) are monospecific genera (Al-Shehbaz and German, 2016; Al-Shehbaz et al., 2004). The genus Baimashania has two species (Baimashania pulvinata Al-Shehbaz and B. wangii Al-Shehbaz) (Al-Shehbaz, 2000). Although new species and genera of Brassicaceae are continually being described, few chromosomal data of related groups have been studied, which hampers comparative research. A statistical data-base has shown that chromosome numbers are generally known from 232 of the 338 (68.6%) genera and 1558 of the 3709 (42.0%) species of Brassicaceae (Koch and Al-Shehbaz, 2004). For Brassi-caceae plants in China, there are 205 species in 74 genera with chromosome reports (statistical data by the authors). Cytological data can provide useful evidence for the systematics and taxonomy of Brassicaceae.

|
| Fig. 1 Photographs of morphological traits and habits of Anzhengxia, Shangrilaia and Baimashania. 1e4: A. yechengnica (Photos by Hong-Liang Chen); 5e8: S. nana (Photos by Ji-Pei Yue); 9e12: B. pulvinata (Photos by Wen-Guang Sun). |
Despite their importance, the relationships among major line-ages in Brassicaceae remain unresolved (Nikolov et al., 2019). To better understand the relationships in the Brassicaceae, we inves-tigated the chromosome number and karyotype asymmetry pat-terns of Anzhengxia, Shangrilaia, and Baimashania, three endemic genera from the Qinghai-Tibet Plateau. To test the hypothesis that polyploidization is the primary driver of speciation in the Qinghai-Tibet Plateau region, we surveyed chromosome numbers and ploidy levels of species in two tribes (Euclidieae and Arabideae) of Brassicaceae that have distributions limited the Qinghai-Tibet Plateau.
2. Materials and methods 2.1. Plant materials and treatmentsAnzhengxia yechengnica, Shangrilaia nana, Baimashania pulvinata plants were collected from northwest and southwest China (Table 1). Voucher specimens have been deposited in the herbari-um of the Kunming Institute of Botany (KUN).
| Taxon | Locality | Coordinates | Altitude/m | Voucher |
| Anzhengxia yechengnica | Yecheng, Xinjiang, China | E77°03'36", N37°11'34" | 2450 | YC-XZ115 |
| Shangrilaia nana | Shangri-La, Yunnan, China | E99°35'13", N27°47'35" | 4408 | CHY-008 |
| Baimashania pulvinata | Deqin, Yunnan, China | E99°01'20", N28°23'21" | 4530 | MS17-157 |
Chromosome numbers were Counted in more than 30 somatic℃ells from the root tips of seedlings of each taxon. Prior to germi-nation, seeds were stored at 4℃ for at least one month. Subse-quently, seeds were germinated in agarose at 24℃. Fresh root tips (about 1-2 cm in length) were excised from the seedlings and pretreated in 0.002 or 0.003 mol/L 8-hydroxyquinoline solution at 24℃ in the dark for 2 h, then fixed in 3:1℃arnoy's solution (ab-solute ethanol: acetic acid, v/v) for at least 24 h at 4℃. The fixed roots were hydrolysed in 1 mol/L HCl at 60℃ for 12 min, and then washed with distilled water, dyed with℃arbolfuchsin and squashed for observation. Karyotype analyses were based on measurements of mitoticemetaphase Chromosomes taken from photographs.
2.2. Karyotype analysisKaryotype analyses were based on at least six mitotic meta-phase cells from each species. Chromosome classifications were made by the standardized nomenclature proposed by Levan et al. (1964). The degree of karyotype asymmetry was estimated with Stebbins's method (Stebbins, 1971). The index of Karyotypic Asymmetry (As.K %) was calculated as As. K% = the total of the longest in a chromosome set/the total of the a chromosome set×100. Asymmetry index (AI) was calculated as AI = CVCL×CVCI/100, where CVCL is a component expressing the relative variation in chromosome length, and CVCI is a component expressing the relative variation in the centromeric index. Kar-yotype parameters were measured using KaryoType software (Altınordu et al., 2016).
2.3. Polyploid frequency within the tribes Euclidieae and ArabideaeTo quantify variation in chromosome number and polyploid frequency within the tribes Euclidieae and Arabideae, we surveyed published reports on species of the tribes (Table 2). For the same purpose, we consulted the network database from Index to Plant Chromosome Numbers (IPCN, http://www.tropicos.org/NameSearch.aspx?projectid=9) (Goldblatt and Lowry, 2011) and Chromosome Counts Database (CCDB, http://ccdb.tau.ac.il/home/) (Rice et al., 2015). For each taxon (including species and subspe-cies), we used the originally published names and proofread the name in The Plant List (TPL, http://www.theplantlist.org/).
| Genus | No. Species | Base | Chromosome No. (n) | Chromosome No. (2n) |
| Counted | Chromosome | |||
| No. (x) | ||||
| Tribe Euclidieae | ||||
| Anzhengxia | 1 | 7 | 7 | 14 |
| Braya | 21 | 6, 7, 8, 9, 10 | 9, 14, 16, 20, 21, 24, 25, 28, | 18, 28, 32, 40, 42, 48, 50, 56, |
| 32, 35, 42, 56 | 64, 70, 84, 112 | |||
| Christolea | 3 | 6, 7 | 6, 7 | 12, 14 |
| Cryptospora | 1 | 7 | 7 | 14 |
| Euclidium | 1 | 7 | 7 | 14 |
| Lachnoloma | 1 | 7 | 7 | 14 |
| Leiospora | 2 | 7 | 7 | 14 |
| Leptaleum | 1 | 7 | 7 | 14 |
| Octoceras | 1 | 7 | 7 | 14 |
| Parrya | 1 | 7 | 7 | 14 |
| Sisymbrium | 1 | 7 | 7 | 14 |
| Solms-laubachia | 8 | 7 | 7, 14 | 14, 28 |
| Streptoloma | 1 | 7, 10 | 7, 40 | 14, 80 |
| Tetracme | 2 | 7 | 7, 14 | 14, 28 |
| Tribe Arabideae | ||||
| Arabis | 104 | 4, 6, 7, 8, 9, 10, | 4, 6, 7, 8, 9, 10, 11, 12, 14, | 8, 12, 14, 16, 17, 18, 20, 22, |
| 11 | 15, 16, 20, 21, 24, 32 | 24, 28, 30, 32, 40, 42, 48, 64 | ||
| Athysanus | 1 | 13 | 13 | 26 |
| Aubrieta | 12 | 8 | 8, 16 | 16, 32 |
| Baimashania | 1 | 8 | 8 | 16 |
| Draba | 204 | 6, 7, 8, 9, 10, 11, | 6, 7, 8, 9, 10, 11, 12, 13, 14, | 12, 14, 16, 18, 20, 22, 24, 26, |
| 12, 13, 15, 19 | 15, 16, 18, 19, 20, 21, 22, 24, | 28, 30, 32, 36, 38, 40, 42, 44, | ||
| 25, 26, 27, 28, 30, 31, 32, 33, | 48, 50, 52, 54, 56, 60, 62, 64, | |||
| 36, 37, 38, 40, 41, 47, 48, 50, | 66, 72, 74, 76, 80, 82, 94, 96, | |||
| 56, 60, 64, 72 | 100, 112, 120, 128, 144 |
We used these same sources to calculate n, 2n and polyploid frequency for species from the tribes Euclidieae (14 taxa, 8 genera) and Arabideae (18 taxa, 2 genera) that are distributed in the Qinghai-Tibet Plateau.
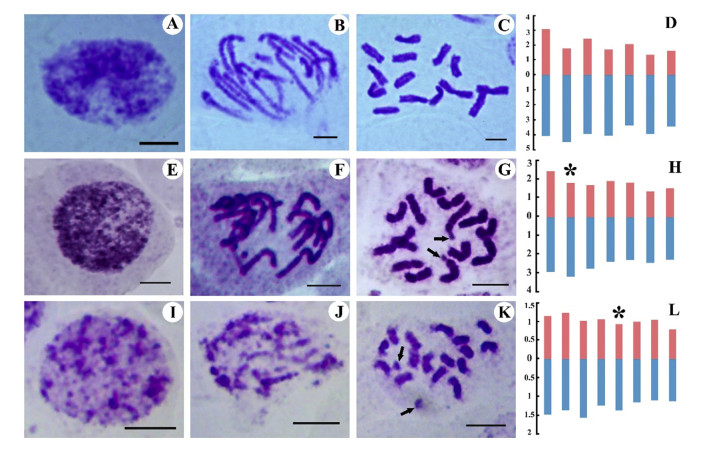
3. Results 3.1. Anzhengxia yechengnica (C. H. An) Al-Shehbaz & D. A. GermanAnzhengxia, a monotypic genus of the tribe Euclidieae, is endemic in the Pamirs and Kunlun Mountains region (Al-Shehbaz and German, 2016). The karyotype formula for A. yechengnica is 2n = 2x = 14 = 6 m + 8sm (Table 3). According to the nomenclature of Tanaka (1971), the interphase nuclei and prophase chromosomes can be categorized as diffuse type (Fig. 2: A, B, C, D). The chromosomes varied in length from 5.05 to 7.11 mm. The proportion of the longest to the shortest chromosome length was 1.41, and the AI = 1.98. The karyotype asymmetry (KA) belongs to Stebbins's-3B (Table 4). This is the first report for the karyotype parameters of A. yechengnica.
| Species | Chromosome pair No. | L (%) | S (%) | L+S(%) | L/S | Chromosome type |
| Anzhengxia yechengnica | 1 | 9.84 | 7.51 | 17.36 | 1.31 | m |
| 2 | 10.92 | 4.33 | 15.26 | 2.52 | sm | |
| 3 | 9.36 | 5.87 | 15.23 | 1.60 | m | |
| 4 | 9.79 | 4.18 | 13.97 | 2.34 | sm | |
| 5 | 8.11 | 4.97 | 13.09 | 1.63 | m | |
| 6 | 9.55 | 3.28 | 12.83 | 2.91 | sm | |
| 7 | 8.33 | 3.95 | 12.28 | 2.11 | sm | |
| Shangrilaia nana | 1 | 9.17 | 8.10 | 17.27 | 1.13 | m |
| 2 | 10.44 | 5.82 | 16.26 | 1.79 | sm (2sat) | |
| 3 | 8.87 | 5.62 | 14.49 | 1.58 | m | |
| 4 | 7.66 | 6.30 | 13.96 | 1.22 | m | |
| 5 | 7.34 | 5.96 | 13.30 | 1.23 | m | |
| 6 | 7.90 | 4.43 | 12.33 | 1.78 | sm | |
| 7 | 7.35 | 5.05 | 12.39 | 1.46 | m | |
| Baimashania pulvinata | 1 | 7.90 | 6.27 | 14.16 | 1.26 | m |
| 2 | 7.29 | 6.66 | 13.94 | 1.09 | m | |
| 3 | 8.38 | 5.52 | 13.90 | 1.52 | m | |
| 4 | 6.66 | 5.73 | 12.38 | 1.16 | m | |
| 5 | 7.29 | 5.01 | 12.31 | 1.45 | m (2sat) | |
| 6 | 6.18 | 5.43 | 11.61 | 1.14 | m | |
| 7 | 5.87 | 5.62 | 11.48 | 1.04 | m | |
| 8 | 5.97 | 4.25 | 10.21 | 1.40 | m | |
| Abbreviations: L (%), relative length of long arm; S (%), relative length of short arm, L + S (%), relative length of total chromosome; L/S, arm ratio; m, metacentric; sm, submetacentric; sat: satellite chromosome. | ||||||
|
| Fig. 2 Mitotic nuclei, metaphase chromosomes, and ideograms of Anzhengxia, Shangrilaia and Baimashania. A-D: A. yechengnica; E-H: S. nana; I-L: B. pulvinata. Scale bar = 5 mm; Red: the relative length of short arm; Blue: the relative length of long arm; arrowheads and asterisks indicate satellite chromosomes. |
| Taxon | Chromosome lengthrange/μm |
Ratio LC/SC | L/μm±SD | S/μm±SD | TCL/μm±SD | CI±SD | >2:1 | As.K% | AI | THL±SD | CVCL | CVCI | MCA | KA | Chromosome No./basicNo./ Polyploidy |
Karyotyp Feormula | Fig. 2 |
| Anzhengxia yechengnica | 5.05-7.11 | 1.41 | 3.88(±0.36) | 2.00(±0.54) | 11.75(±0.67) | 33.37(±5.90) | 0.57 | 66.01 | 1.98 | 5.88(±0.67) | 11.44 | 17.34 | 32.33 | 3A | 14/7/2x | 6m+8sm | A, B, C, D |
| Shangrilaia nana | 3.78-5.34 | 1.41 | 2.57(±0.32) | 1.80(±0.31) | 8.75(±0.54) | 41.17(±3.77) | 0 | 58.81 | 1.17 | 4.37(±0.54) | 12.43 | 9.40 | 17.85 | 1A | 14/7/2x | 10m+4sm(2sat) | E, F, G, H |
| Baimashania pulvinata | 1.90-2.61 | 1.37 | 1.28(±0.32) | 1.03(±0.12) | 4.62(±0.24) | 44.48(±0.32) | 0 | 55.61 | 0.76 | 2.31(±0.24) | 10.39 | 7.35 | 11.11 | 1A | 16/8/2x | 12m(2sat)+4sm | I, J, K, L |
| Abbreviations: LC/SC, the proportion of the longest chromosome length to the shortest chromosome length; L, long arm length (μm); S: short arm length (μm); TCL, mean length of chromosome (μm); CI, mean centromeric index; As.K%, index of karyotypic asymmetry (Arano, 1963); AI, asymmetry index (Paszko, 2006); THL, Total haploid (monoploid) length of chromosome set (μm); CVCL, coefficient of variation of chromosome length (Paszko, 2006); CVCI, coefficient of variation of the centromeric index (Paszko, 2006); MCA, mean centromeric asymmetry (Peruzzi and Eroğlu, 2013); KA, karyotype asymmetry (Stebbins, 1971); m, metacentric; sm, submetacentric; sat, satellite chromosome; SD, standard deviation. | |||||||||||||||||
Shangrilaia is a monotypic genus of tribe Euclidieae with S. nana and exhibits a typical distribution in the Hengduan Mountains re-gion, SW China (Al-Shehbaz et al., 2004). S. nana grows naturally in alpine gravel meadows or extremely weathered gravelly slopes at altitudes mostly above 4200 m. The karyotype of S. nana is 2n = 2x = 14 = 10 m + 4sm (2sat) (Table 3). According to the nomenclature of Tanaka (1971), the interphase nuclei and prophase chromosomes can be categorized as diffuse type (Fig. 2: E, F, G, H). The second chromosome pair of diploid S. nana has a satellite chromosome on the short arm (Table 3). The chromosomes varied in length from 3.78 to 5.34 mm. The ratio of the longest to the shortest chromosome length was 1.41, and the AI = 1.17; KA is Stebbins's-1A (Table 4). This is the first report for the karyotype parameters of S. nana.
3.3. Baimashania pulvinata Al-ShehbazBaimashania is a genus of the Brassicaceae with two identified species (B. pulvinata and B. wangii) (Al-Shehbaz, 2000). B. pulvinata is restricted mainly to alpine scree or weathered rocks in the Hengduan Mountains region at altitudes ranging from 4100 to 4600 m (Xu et al., 2014). This species is morphologically similar to plants in three genera (Solms-laubachia Muschler, Leiospora (C.A. Meyer) Dvorak, Pycnoplinthopsis Jafri). The karyotype formula of B. pulvinata is 2n = 2x = 16 = 12 m (2sat) + 4sm (Table 3). Ac-cording to the nomenclature of Tanaka (1971), the interphase nuclei and prophase chromosomes can be categorized as simple type (Fig. 2: I, J, K, L). The fifth chromosome pair in the diploid has one satellite on the short arm (Table 3). The basic chromosome number of Baimashania was x = 8 and the somatic cells of the sampled B. pulvinata were diploid. The chromosomes varied in length from 1.90 to 2.61 mm. The ratio of the longest to the shortest chromo-some was 1.37, the AI = 0.76. KA is Stebbins's-1A (Table 4). This is the first report of the karyotype of B. pulvinata.
3.4. Summary of chromosome numbers for the tribes Euclidieae and ArabideaeChromosome number for 1931 taxa of Brassicaceae in 258 genera have been reported (Goldblatt and Lowry, 2011; Rice et al., 2015; Warwick and Al-Shehbaz, 2006). In the Brassicaceae tribe Euclidieae, chromosome numbers have been reported for 45 taxa in 14 genera. Chromosome number varies greatly in the tribe Eucli-dieae, with at least 16 different chromosome numbers, ranging from 2n = 12 to 2n = 112 (Table 2). Over 93.33% of taxa in tribe Euclidieae have the basic chromosome number x = 7.
Chromosome numbers have been reported for 322 taxa from 5 genera of tribe Arabideae. In the tribe Arabideae, at least 39 different chromosome numbers have been found, ranging from 2n = 8 to 2n = 144 (Table 2). Over 86.96% of taxa in the tribe Arabideae have the basic chromosome number x = 8.
Our survey of chromosome numbers and ploidy levels in tribes Euclidieae and Arabideae (367 species in 19 genera) showed that ploidy levels varied from 2x to 24x (Table 2).
We also surveyed chromosome number and ploidy levels for species that belong to the tribes Euclidieae and Arabideae but whose distribution is restricted to the Qinghai-Tibet Plateau region (Table 5). We found that the polyploid frequency of these species is about 36.11%.
| Taxon | Chromosome No. (n) | Chromosome No. (2n) | Polyploidy |
| Tribe Euclidieae | |||
| Braya rosea | 14 | 28 | 4x |
| Christolea crassifolia | 7 | 14 | 2x |
| Euclidium syriacum | 7 | 14 | 2x |
| Leiospora bellidifolia | 7 | 14 | 2x |
| Leptaleum filifolium | 7 | 14 | 2x |
| Neotorularia torulosa | 7 | 14 | 2x |
| Solms-laubachia eurycarpa | 7 | 14 | 2x |
| Solms-laubachia linearifolia | 7 | 14 | 2x |
| Solms-laubachia minor | 7 | 14 | 2x |
| Solms-laubachia pulcherrima | 7 | 14 | 2x |
| Solms-laubachia retropilosa | 7, 14 | 14, 28 | 2x, 4x |
| Solms-laubachia xerophyta | 7 | 14 | 2x |
| Tetracme pamirica | 14 | 28 | 4x |
| Tetracme quadricornis | 7 | 14 | 2x |
| Tribe Arabideae | |||
| Arabis amplexicaulis | 8, 16 | 16, 32 | 2x, 4x |
| Arabis hirsuta | 16 | 32 | 2x |
| Arabis pterosperma | 8 | 16 | 2x |
| Arabis tibetica | 8 | 16 | 2x |
| Draba alajica | 20 | 40 | 4x |
| Draba alpina | 31, 32, 33, 37, 40, 56, 60 | 62, 64, 66, 74, 80, 112, 120, 64-66 | 2x |
| Draba altaica | 8, 9 | 16, 18 | 2x, 4x |
| Draba cana | 16 | 32 | 4x |
| Draba draboides | 21 | 42 | 6x |
| Draba eriopoda | 8 | 16 | 2x |
| Draba glacialis | 8 | 16 | 2x |
| Draba lanceolata | 16, 24 | 24, 32 | 3x, 4x |
| Draba melanopus | 16 | 32 | 4x |
| Draba nemorosa | 8 | 16 | 2x |
| Draba nuda | 8 | 16 | 2x |
| Draba olgae | 6 | 12 | 2x |
| Draba oreades | 20 | 40 | 4x |
| Draba subamplexicaulis | 24 | 48 | 6x |
Our study presents an integral survey on the karyological vari-ation of all currently recognized members of the tribes Euclidieae and Arabideae. In this study, we found that A. yechengnica and S. nana have the same chromosome number, x = 7, and that this number is the same as the basic chromosome number of plants in tribe Euclidieae. This finding supports the placement of the genera Anzhengxia and Shangrilaia in tribe Euclidieae (Chen et al., 2018). In addition, we report the first formulation of the karyotype of B. pulvinata, which has a basic chromosome number x = 8 and diploid number 2n = 16.
We formulated the karyotype of A. yechengnica as 2n = 2x = 14 = 6 m + 8sm. This is the first report of karyotype parameters for A. yechengnica. Traditionally, A. yechengnica has been placed in Microsisymbrium O. E. Schulz as M. yechengicum Z.X. An (An, 1981); however, M. yechengicum was transferred to Sisymbriopsis Botsch. & Tzvelev as S. yechengicum Z. X. An (1981) by Al-Shehbaz et al. (1999). Recent molecular phylogenetic study and morphological characters suggested that S. yechengnica is a monotypic genus of the tribe Euclidieae and should be treated as A. yechengnica (Al-Shehbaz and German, 2016). The occurrence of 2n = 2x = 14 in A. yechengnica is similar to the karyotype of Sisymbriopsis mollipila (Maximowicz) Botschantzev (Ren et al., 2008), which has the same chromosome number as most spe-cies of the tribe Euclidieae.
The genus Shangrilaia of tribe Euclidieae is monotypic, con-sisting of S. nana, which is typically distributed in northwest Yunnan (Shangri-La County) (Al-Shehbaz et al., 2004). In this study, we found that the karyotype of S. nana is 2n = 2x = 14 = 10 m + 4sm (2sat). In addition, we discovered that the second chromosome pair of diploid S. nana has a satellite chromosome on the short arm. Our results show that S. nana has the same chromosome count (x = 7) as Solms-laubachia (Yue et al., 2003, 2004), which is the most frequent chromosome count of the tribe Euclidieae. S. nana is also morphologically similar to Solms-laubachia in flowering pattern, petal color, and seed characteris-tics. However, the fruit of these two species differ; Solms-laubachia fruit are silique, whereas Shangrilaia fruit are silicle (Fig. 1) (Al-Shehbaz et al., 2004).
Baimashania is a genus within Brassicaceae with two identi-fied species (B. pulvinata and B. wangii) (Al-Shehbaz, 2000). Morphologically, these two species are closely related to the genera Solms-laubachia, Leiospora, and Pycnoplinthopsis. In the "Flora Reipublicae Popularis Sinicae", B. pulvinata was illustrated as Solms-laubachia ciliaris (Bur. et Franch.) Botsch. However, morphological studies established the new genus of Baimashania (Al-Shehbaz, 2000). Our study reveals that the basic chromosome number of B. pulvinata is x = 8, and chromosome pair five of the diploid has one satellite on the short arm. This finding is consistent with the base chromosome number in tribe Arabideae. Furthermore, the basic chromosome number and karyotype of B. pulvinata differs from Solms-laubachia (Yue et al., 2003, 2004) and is distinct from the base number x = 7 in tribe Euclidieae. Combined with previous studies (Yue et al., 2004; Yue et al., 2003), our findings support the placement of the genus Baima-shania in tribe Arabideae.
4.2. Polyploid frequency in the tribes Euclidieae and ArabideaePolyploidization is a key factor of plant diversity and speciation (Ehrendorfer, 1980; Lewis, 1980; Ramsey and Schemske, 2002; Stebbins, 1971). Our survey of chromosome numbers and ploidy levels in tribes Euclidieae and Arabideae (367 species in 19 genera) showed that ploidy levels varied from 2x to 24x (Table 2). In some species complexes, diploids are aneuploid, which may result in future hybridization and polyploidy and the creation of complex patterns of chromosome numbers. We have confirmed the exis-tence of different cytotypes of the same ploidy level. More evidence is required of cytogenetics and genome size from related groups.
We found that the polyploid frequency in Euclidieae and Ara-bideae taxa distributed exclusively in the Qinghai-Tibet Plateau was about 36.11%. Although more taxa should be examined, this finding does not support the hypothesis that the polyploidy increases adaptability to extreme environments encountered in the Qinghai-Tibet Plateau.
Authors' contributionH.S. and Z-M.L. designed the study. W-G.S., H.-X.W. and R.W. performed the experiments. W-G.S. analyzed the data. W-G.S. wrote the first draft of the manuscript. All authors read and approved the final manuscript.
Declaration of Competing InterestThe authors declare that no potential conflict of interest.
AcknowledgmentsWe are grateful to Dr Hong-Liang Chen and Dr Ji-Pei Yue for providing the seed materials and taking the photographs of plants. This work was supported by grants from the National Natural Sci-ence Foundation of China (grant numbers 31960046 and 31670206 to Z.-M.L.), Strategic Priority Research Program of Chinese Academy of Sciences, China (grant number XDA 20050203 to H.S.), and Key Projects of the Joint Fund of the National Natural Science Founda-tion of China (grant number U1802232 to H.S.). We also acknowl-edge TopEdit LLC for the linguistic editing and proofreading during the preparation of this manuscript.
Al-Shehbaz I.A, 2020. Baimashania (Brassicaceae), a new genus from China. Novon, 10: 320-322. |
Al-Shehbaz I.A, An Z.X., Yang G., 1999. A revision of Sisymbriopsis (Brassicaceae). Novon, 14: 271-274. |
Al-Shehbaz I.A, German D.A., 2016. Three new genera in the tribe Euclidieae(Brassicaceae). Novon, 25: 12-17. DOI:10.3417/2016015 |
Al-Shehbaz I.A., Yue J.P., Sun H., 2004. Shangrilaia (Brassicaceae), a new genus from China. Novon, 14: 271-274. |
Altınordu F, Peruzzi L., Yu Y., He X.J., 2016. A tool for the analysis of chromosomes: KaryoType. Taxon, 65: 586-592. DOI:10.12705/653.9 |
An C.H., 1981. New materials for Chinese Cruciferae. Bull. Bot. Res, 1: 97-107. |
Appel, O., Al-Shehbaz, I., 2003. Cruciferae. In: Kubitzki, K., Bayer, C. (Eds.), The Families and Genera of Vascular Plants 5. Springer, Berlin, pp. 75-174.
|
Arano H., 1963. Cytological studies in subfamily Carduoideae (Compositae) of Japan IX. The karyotype analysis and phylogenic considerations on Pertya and Ainsliaea. Bot. Mag, 76: 32-39. |
Chen, H.L., Al-Shehbaz, I.A., Yue, J.P., Sun, H., 2018. New insights into the taxonomy of tribe Euclidieae (Brassicaceae), evidence from nrITS sequence data. PhytoKeys 100, 125-139.
|
Ehrendorfer F., 1980. Polyploidy and distribution. In: Lewis W.H. (Ed.), Polyploidy:Biological Relevance. Washington University, Washington, D.C., pp: 45-60. |
Goldblatt P., Lowry P.P., 2011. The index to plant chromosome numbers (IPCN):three decades of publication by the Missouri Botanical Garden come to an end. Ann. Mo. Bot. Gard, 98: 226-228. DOI:10.3417/2011027 |
Guerra M., 2008. Chromosome numbers in plant cytotaxonomy: concepts and implications. Cytogenet. Genome Res, 120: 339-350. DOI:10.1159/000121083 |
Hanelt P., 1966. Polyploidie-Frequenz und geographische Verbreitung bei höheren Pflanzen. Biologische Rundschau. vol, 4(pp): 183-196. |
Kiefer M., Schmickl R., German D.A., Mand#225;ková T. Lysak M.A., Al-Shehbaz I.A., Franzke A., Mummenhoff K., Stamatakis A., Koch M.A., 2014. BrassiBase:introduction to a novel knowledge database on Brassicaceae evolution. Plant Cell Physiol, 55: e3. DOI:10.1093/pcp/pct158 |
Koch M., Al-Shehbaz I.A., 2004. Taxonomic and phylogenetic evaluation of the American pThlaspiq species: identity and relationship to the Eurasian genus Noccaea (Brassicaceae). Syst. Bot, 29: 375-384. DOI:10.1600/036364404774195566 |
Levan, A., Fredga, K., Sandberg, A.A., 1964. Nomenclature for centromeric position on chromosomes. Hereditas 52, 201-220.
|
Lewis W.H., 1980. Polyploidy in Angiosperms: Dicotyledons, Polyploidy. Springer, pp: 241-268. |
Löve, Á., Löve, D., 1967. Polyploidy and altitude: Mt. Washington. Biol. Zentralblatt 86, 307-312.
|
Löve, Á., Löve, D., 1975. Cytotaxonomical Atlas of the Arctic Flora. J. Cramer, Vaduz.
|
Nikolov L.A., Shushkov P., Nevado B., Gan X.C., Al-Shehbaz I.A., Filatov D., Bailey C.D., Tsiantis M., 2019. Resolving the backbone of the Brassicaceae phylogeny for investigating trait diversity. New Phytol, 222: 1638-1651. DOI:10.1111/nph.15732 |
Paszko B., 2006. A critical review and a new proposal of karyotype asymmetry indices. Plant Systemat. Evol, 258: 39-48. DOI:10.1007/s00606-005-0389-2 |
Peruzzi L., Ero#287;lu H.E., 2013. Karyotype asymmetry: again how to measure and what to measure? Comp. Cytogenet, 7: 1-9. DOI:10.3897/compcytogen.v7i1.4431 |
Ramsey J., Schemske D.W., 2002. Neopolyploidy in flowering plants. Annu. Rev. Ecol. Systemat, 33: 589-639. DOI:10.1146/annurev.ecolsys.33.010802.150437 |
Ren H.Y., Tuo Z.Y., Wumaierxiati T., Ling Z.G., 2008. Chromosome numbers of 8 species of Cruciferae in Xinjiang. J. Xinjiang Agric. Univ, 31: 8-11. |
Rice A., Glick L., Abadi S., Einhorn M., Kopelman N.M., Salman-Minkov A., Mayzel J., Chay O., Mayrose I., 2015. The Chromosome Counts Database (CCDB)-a community resource of plant chromosome numbers. New Phytol, 206: 19-26. DOI:10.1111/nph.13191 |
Soltis P.S., Soltis D.E., 2000. The role of genetic and genomic attributes in the success of polyploids. Proc. Natl. Acad. Sci. U.S.A, 97: 7051-7057. DOI:10.1073/pnas.97.13.7051 |
Spicer R.A., 2017. Tibet, the Himalaya, Asian monsoons and biodiversityeIn what ways are they related?. Plant Divers, 39: 233-244. DOI:10.1016/j.pld.2017.09.001 |
Stebbins, G.L., 1971. Chromosomal Evolution in Higher Plants. Addison Wesley, New York.
|
Sun H., Zhang J.W., Deng T., Boufford D.E., 2017. Origins and evolution of plant diversity in the Hengduan Mountains, China. Plant Divers, 39: 161-166. DOI:10.1016/j.pld.2017.09.004 |
Sun W.G., Sun H., Li Z.M., 2019. Chromosome data mining and its application in plant diversity research. Plant Sci. J, 37: 260-269. |
Tanaka R., 1971. Types of resting nuclei in Orchidaceae. Bot. Mag, 84: 118-122. DOI:10.15281/jplantres1887.84.118 |
Warwick S.I., Al-Shehbaz I.A., 2006. Brassicaceae: chromosome number index and database on CD-Rom. Plant Systemat. Evol, 259: 237-248. DOI:10.1007/s00606-006-0421-1 |
Xu, B., Li, Z.M., Sun, H., 2014. Seed Plants of the Alpine Subnival Belt from the Hengduan Mountains, SW China. Science Press, Beijing.
|
Yue J.P., Gu Z.J., Al-Shehbaz I.A., Sun H., 2004. Cytological studies on the SinoHimalayan endemic Solms-laubachia (Brassicaceae) and two related genera. Bot. J. Linn. Soc, 145: 77-86. DOI:10.1111/j.1095-8339.2003.00268.x |
Yue J.P., Sun H., Al-Shehbaz I.A., Gu Z.J., 2003. Cytological studies of five Chinese species of Solms-laubachia (Brassicaceae). Harv. Pap. Bot: 467-473. |
Zhou, T.Y., Cheo, T.Y., Lu, L.L., Lou, L.L., Yang, G., Al-Shehbaz, I.A., 2001. Brassicaceae. In: Wu, C.Y., Raven, P.H. (Eds.), Flora of China. Science Press, Beijing, pp. 1-193. St. Louis: Missouri Botanical Garden Press.
|



