b. Kunming College of Life Science, University of Chinese Academy of Sciences, Kunming, Yunnan, 650204, China;
c. College of Life Sciences, Southwest Forestry University, Kunming, Yunnan, 650224, China
Fructose-1, 6-bisphosphatases (FBPase) participate in gluconeogenesis and the Calvin cycle during photosynthesis by catalyzing the irreversible hydrolysis of fructose-1, 6-bisphosphate into fructose-6-phosphate and inorganic phosphate. FBPases are members of the large superfamily of lithium-sensitive phosphatases, which consists of three families of inositol phosphatases and FBPases. Five types of FBPases have been found in prokaryotes, while only one type of FBPase, homologous to prokaryotic FBPaseI, has been found in eukaryotes (Donahue et al., 2000; Rashid et al., 2002; Verhees et al., 2003; Brown et al., 2009). In photosynthetic eukaryotes (algae and plants), two FBPase isoforms have previously been identified. More recently, the discovery of a novel FBPase isoform has raised questions about the function, distribution, and evolutionary relationships of FBPases.
The structure and function of two FBPase isoforms in photosynthetic eukaryotes have been well characterized. The first isoform, cyFBPase, is a cytosol-localized enzyme that participates in gluconeogenesis and sucrose synthesis. cyFBPase is similar to analogs in heterotrophic eukaryotes that also play a role in cytosolic gluconeogenesis (Cseke and Buchanan, 1986). The second isoform, cpFBPase (hereafter, cpFBPase1), is present in the stroma of chloroplasts, where it is one of the key regulatory components of the Calvin cycle. cpFBPase, but not the cytosolic isoform, contains an 20-30-aa insertion sequence called the 'loop 170' that acts as a regulatory redox domain. This domain generally contains three conserved cysteines (Cys) that are able to form a regulatory disulphide and confer exclusive redox features to the chloroplast isoform (Chiadmi et al., 1999; Chueca et al., 2002). cyFBPase and cpFBPase1 differ strikingly in how their enzymatic activity is regulated. The activity of cpFBPase1 is stimulated by lightmodulated reduction of essential disulphide groups via the ferredoxin-thioredoxin system (Buchanan, 1980), whereas the activity of cyFBPase is similar to that of yeast and mammals, in which metabolic affectors such as adenosine monophosphate (AMP) act as inhibitors (Zimmermann et al., 1978). However, the regulation and physiological function of the newly identified FBPase remains unclear.
This novel isoform, named cpFBPaseII (Serrato et al., 2009) or chloroplastic FBPaseII (hereafter FBPase2) (Ogawa et al., 2015), differs from cpFBPase1, because it lacks a regulatory redox domain. Although previous studies suggested that cpFBPase2 may play a role in development, metabolism, or stress response in multicellular organisms (Serrato et al., 2009), this enzyme has subsequently been identified in a unicellular phytoflagellate, Euglena gracilis, where it is called EgFBPaseII. EgFBPaseII's high oxidation tolerance has prompted researchers to hypothesize that cpFBPase2 is required during oxidative stress (Ogawa et al., 2015).
Early phylogenetic analysis concluded that although cpFBPase1 is localized and functions in chloroplasts, it was not inherited directly from the chloroplast ancestors, i.e., cyanobacteria, but probably originated through duplication of the cytosolic isoform, cyFBPase (Martin et al., 1996). Although cpFBPase2 has been hypothesized to be restricted to land plants, no studies have examined its distribution in the key lineages of photosynthetic eukaryotes.
The overall goal of this study was to understand the evolutionary origins and physiological functions of chloroplast FBPases. Specifically, we asked two questions:(1) is the cpFBPase2 isoform restricted to land plants? (2) Do FBPases belong to a monophyletic group? To answer these questions, we identified FBPases based on sequence homology in 33 photosynthetic eukaryotes, which included groups with primary plastids and groups with secondary plastids. We classified FBPases as one of the three isoforms by using sequence data to predict subcellular localization and the presence/ absence of 'loop 170.' Finally, we analyzed the phylogenetic relationship of FBPases from these organisms. Our study indicates the following:(1) in addition to the early-discovered cyFBPase and cpFBPase, the later-identified cpFBPase isoform also exists ubiquitously in photosynthetic eukaryotes; (2) the formation of the three FBPase isoforms was due to the gene duplication of cyFBPase to produce the later-identified cpFBPase, which was then duplicated to produce the early-discovered cpFBPase; and (3) the two cpFBPases most likely coexist in chloroplasts in order to cope with different redox conditions during photosynthesis to ensure its continued action under different light conditions.
2. Materials & methods 2.1. Identification of FBPases in diverse photosynthetic eukaryotesThe sequence of the cpFBPase2 isoform (Fragaria × ananassa cpFBPaseII, accession number:ABW38331) was used as a query to search for FBPases from diverse photosynthetic eukaryotes by utilizing Blast software (Altschul et al., 1990) (E < 0.001) (detailed species information is provided in Table 1). The hits obtained were further examined with a Pfam search (http://pfam.sanger.ac.uk/). Only the candidate sequences with a typical FBPase domain (FBPaseI domain, accession number:PF00316.20) were reblasted against the UniProt database to determine whether their top hits were FBPases. The sequences that were retained during this process were identified as FBPases; their unique identifiers are provided in Supplemental Data S1.
| Species | cyFBPase | cpFBPase1 | cpFBPase2 | ||
| Primary plastid lineages | Land plants | Marchantia polymorpha | + | + | + |
| Physcomitrella patens | + | + | + | ||
| Selaginella moellendorffii | + | + | + | ||
| Arabidopsis thaliana | + | + | + | ||
| Fragaria x ananassa | + | + | + | ||
| Chlorophyta | Auxenochlorella protothecoides | + | + | -? | |
| Bathycoccus prasinos | + | + | + | ||
| Chlamydomonas reinhardtii | - | + | - | ||
| Chlorella sorokiniana DOE 1412 / UTEX 1230 / str. 1228 | + | + | - | ||
| Chlorella variabilis NC64A | + | + | -? | ||
| Chromochloris zofingiensis | - | + | + | ||
| Dunaliella salina | + | + | -? | ||
| Micromonas pusilla CCMP 1545 | + | + | + | ||
| Micromonas commoda RCC 299 | + | + | + | ||
| Monoraphidium neglectum | + | + | -? | ||
| Ostreococcus lucimarinus | + | + | + | ||
| Ostreococcus tauri | + | + | + | ||
| Ostreococcus sp. RCC809 | + | + | + | ||
| Picochlorum sp. SENEW3 | + | + | -? | ||
| Volvox carteri | - | + | - | ||
| Rhodophyta | Chondrus crispus | + | + | -? | |
| Cyanidioschyzon merolae | + | + | - | ||
| Galdieria sulphuraria | + | + | -? | ||
| Porphyridium purpureum | + | + | + | ||
| Porphyra umbilicalis | + | + | -? | ||
| Glaucophyta | Cyanophora paradoxa | + | + | -? | |
| Secondary plastid lineages | Stramenopiles | Thalassiosira oceanica | + | + | + |
| Phaeodactylum tricornutum | + | + | + | ||
| Ectocarpus siliculosus | + | + | + | ||
| Rhizaria | Bigelowiella natans | + | + | + | |
| Discoba | Euglena gracilis | + | + | + | |
| Note: The symbols '+' and '-' indicate the presence and absence of the enzyme in the corresponding species; the symbol "?" indicates that the absence of a certain FBPase is uncertain due to incomplete genome sequences, which may affect the identification of FBPases. | |||||
To further classify the obtained FBPases into the three isoforms, the following steps were performed. First, subcellular localization prediction for each identified FBPase was performed using the TargetP webserver (http://www.cbs.dtu.dk/services/TargetP) (Emanuelsson et al., 2007) to distinguish cyFBPases from cpFBPases. Second, considering that the regulatory domain (called 'loop 170') that contains three conserved cysteines is the key feature distinguishing the early-discovered cpFBPase from the lateridentified cpFBPase, multiple sequence alignments of the identified FBPases were conducted using the Expresso mode of T-Coffee (Armougom et al., 2006; Di Tommaso et al., 2011; Notredame et al., 2000), to distinguish these two types of cpFBPases. Third, phylogenetic analysis was also used to help identify the three isoforms of FBPases (for analytical details, please see the following section).
2.2. Phylogenetic analysisTo classify the identified FBPases and their evolutionary relationships, phylogenetic analysis was carried out. The aboveidentified FBPase orthologs, together with outgroups (bacterial FBPases with the UniProt identifiers A1VD23 and Q3MCI4), were aligned with the Expresso mode of T-Coffee (Armougom et al., 2006; Di Tommaso et al., 2011; Notredame et al., 2000), a structure-guided multiple sequence alignment mode. The alignment was visually inspected to remove ambiguously aligned regions and gaps (for the final multiple sequence alignment results, please see Supplemental Data S2). The optimal amino acid substitution model was selected using ProtTest version 3.4 (Darriba et al., 2011). Finally, the LG model with a gamma distribution was selected as the best substitution model according to the Akaike information criterion (AIC), corrected AIC (AICc) and Bayesian information criterion (BIC).
Phylogenetic analysis was carried out using both maximumlikelihood (ML) and Bayesian methods, mainly based on the method described in a previous study (Ye et al., 2017). The main procedures were as follows:1) RAxML 7.7.9 (Stamatakis, 2006) was used for rapid bootstrap support (Rbs), and 2) MrBayes 3.2.6 (Ronquist et al., 2012) was used to perform Bayesian analyses. The burn-in fraction was set to 0.25, and the convergence of the chains was assessed with the average standard deviation of split frequencies (ASDSF < 0.01). The 50% majority-rule consensus tree was determined to calculate the posterior probabilities for each node.
3. Results 3.1. The ubiquitous distribution of cpFBPase2 and the pervasive coexistence of chloroplastic FBPases in photosynthetic eukaryotesIn this study, we searched for the presence of FBPases in the genomes of 33 species representative of the main groups of photosynthetic eukaryotes, including those with primary plastids (Glaucophyta, Rhodophyta, Chlorophyta, and land plants) and secondary plastids (Stramenopiles, Rhizaria, and Discoba). The FBPases we identified from photosynthetic eukaryote genomes can be divided into three types based on predictions of subcellular localization and the presence/absence of 'loop 170' (Fig. 1; Supplemental Data S1, S3 and Supplemental Fig. S1, S2). The first type of FBPase is predicted to be localized to the cytosol and lacks the insertion loop sequenced called 'loop 170' as well as the conserved Cys residues within the loop. The second type of FBPase is predicted to be localized in the chloroplast and possesses 'loop 170' as well as two (C95 and C162) or three (C95, C162 and C176) of the conserved Cys residues within the loop (see Fig. 1). These FBPases correspond to those previously referred to as cpFBPaseI and are hereafter referred to as cpFBPase1. The third type of FBPase is predicted to be localized to the chloroplast and possesses 'loop 170.' However, this type of FBPase lacks the conserved Cys residues in 'loop 170.' These FBPases correspond to those previously referred to as cpFBPaseII and are hereafter referred to as cpFBPase2s.
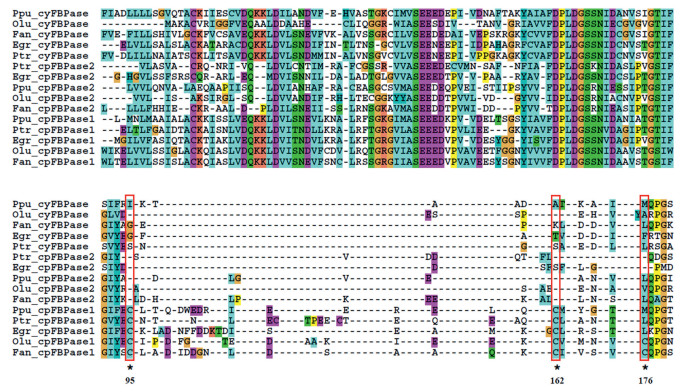
|
| Fig. 1 Partial sequence alignment of cyFBPases and cpFBPases from some investigated photosynthetic eukaryotes, mainly focusing on the region of the redox regulatory domain. Asterisks indicate conserved cysteine residues potentially associated with regulatory disulfide bonds. The red open box indicates the featured amino acid residuals that might be involved in redox regulation in different FBPase types. The sequence alignment results of the cyFBPases and cpFBPases from all the investigated photosynthetic eukaryotes are provided in Supplementary Data S1. The FBPases and their unique identifiers, in parentheses, are Ppu_cyFBPase (contig_3402.5), Olu_cyFBPase (XP_001419675.1), Fan_cyFBPase (ABW38332.1), Egr_cyFBPase (ABF68600.1), Ptr_cyFBPase (23247), Ptr_cpFBPase2 (31994), Egr_cpFBPase2 (BAU20291.1), Ppu_cpFBPase2 (contig_3490.10), Olu_cpFBPase2 (XP_001417955.1), Fan_cpFBPase2 (ABW38331.1), Ppu_cpFBPase1 (contig_3540.6), Ptr_cpFBPase1 (2793), Egr_cpFBPase1 (ABF68597.1), Olu_cpFBPase1 (XP_001422674.1), Fan_cpFBPase1 (ABW38330.1). |
All three types of FBPases are widely distributed in photosynthetic eukaryotes (Table 1). cpFBPase1 and cpFBPase2 coexist in the chloroplasts of most photosynthetic eukaryotes. Specifically, the cytosolic FBPase and cpFBPase1 were detected in almost all photosynthetic eukaryotes. cpFBPase2 was detected in all lineages of photosynthetic eukaryotes investigated, although not in every species. For example, cpFBPase2 was detected in all land plants, in nearly half the green algae (e.g., Bathycoccus prasinos, Ostreococcus spp), in one red algae species (Porphyridium purpureum), in diatoms and filamentous brown algae (e.g., Thalassiosira oceanica and Ectocarpus siliculosus) as well as in unicellular chlorarachniophyte algae (Bigelowiella natans) and the previously reported single cell flagellate eukaryote E. gracilis.
For some green algae (e.g., Auxenochlorella protothecoides, Chlorella variabilis, Dunaliella salina, Monoraphidium neglectum and Picochlorum sp. SENEW3), red algae (e.g., Chondrus crispus, Galdieria sulphuraria and Porphyra umbilicalis), and Glaucophyta, incomplete and/or poor-quality genomes may have inhibited our detection of cpFBPase2 (Blanc et al., 2010; Bogen et al., 2013; Brawley et al., 2017; Collen et al., 2013; Foflonker et al., 2015; Gao et al., 2014; Polle et al., 2017; Schoenknecht et al., 2013).
3.2. The phylogenetic relationships among the three FBPases and the evolutionary divergence of the two cpFBPases in photosynthetic eukaryotesOur phylogenetic analyses of FBPases in diverse photosynthetic eukaryotes revealed that each FBPase type (cytosolic FBPase, cpFBPase1, and cpFBPase2) consistently formed monophyletic groups (Fig. 2). The two monophyletic chloroplast FBPase clades, namely, cpFBPase1 and cpFBPase2, form sister clades. The cyFBPases form an outgroup to the two chloroplastic FBPase isoforms. Our ML and MrBayes tree both showed that, within the cpFBPase1 clade, organisms belonging to land plants and Chlorophyta cluster together to form a monophylic Chloroplastida group, whereas organisms belonging to Rhodophyta from a separate monophyly.
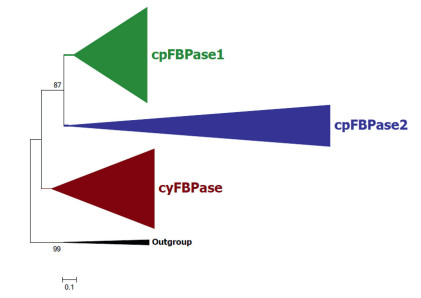
|
| Fig. 2 Phylogenetic analyses of the three types of FBPase isoforms in diverse photosynthetic eukaryotes by ML. cpFBPase1s are in green, cpFBPase2s are in blue, and cyFBPases are in maroon. The tree is rooted using the bacterial FBPaseIs as outgroups. The expanded phylogenetic trees constructed by both ML and MrBayes methods can be found in Supplemental Figures S1 and S2. |
In this study, we found that three types of FBPases-cytoslic FBPase, cpFBPase1, and cpFBPase2-are ubiquitous in almost every lineage of photosynthetic eukaryote. We also found that cpFBPase2s from diverse photosynthetic eukaryotes cluster to form a monophyletic group, indicating that these genes have a single origin.
Our identification of cpFBPase2 in all photosynthetic eukaryotic lineages contrasts with previous hypotheses that cpFBPase2 is restricted to land plants and that it emerged later in the evolution of photosynthetic organisms (Serrato et al., 2009). This difference may be explained by the paucity of whole-genome sequence data available to previous studies. In contrast, our study used genome data from 33 representative photosynthetic eukaryotes.
Although cpFBPase2 was detected in all photosynthetic eukaryotic lineages examined, it was not detected in each genome database of the investigated species of some lineages. There are two possible explanations for this. First, the absence of cpFBPase2 in a specific genome might be due to the incompleteness of genome sequences. For algae whose genome data are incomplete (e.g., A. protothecoides, C. variabilis, D. salina, M. neglectum, Picochlorum sp. SENEW3, C. crispus, G. sulphuraria and P. umbilicalis), cpFBPase2 will likely be detected in these species once their genome sequences are complete. Second, the cpFBPase2 gene may have been secondarily lost in some species. For example, the genome of Chlamydomonas reinhardtii, which is an important model organism, has been completely assembled and annotated with high-quality data at the chromosome level (Merchant et al., 2007); thus, the absence of cpFBPase2 in this organism cannot be attributed to the genome incompleteness. C. reinhardtii cytosolic FBPase has been secondarily lost (Teich et al., 2007); thus, the absence of cpFBPase2 in this organism may also be due to secondary loss. This explanation is significantly strengthened by our phylogenetic analysis (see below).
Previous studies showed that chloroplastic FBPase (cpFBPase1) arose from cytosolic FBPase (cyFBPase) through gene duplication (Martin et al., 1996). Our phylogenetic analyses confirmed this finding by showing that cyFBPase forms an outgroup to the two types of cpFBPases (cpFBPase1 and cpFBPase2). Furthermore, our phylogenetic analyses indicate that cpFBPase1 and cpFBPase2 form sister groups, suggesting that these genes were products of a later gene duplication event.
Although our tree provides strong evidence that the universal common ancestor of the FBPases in eukaryotic photosynthesizers underwent gene duplication to give rise to the ancestral cpFBPase and cyFBPase, the branch lengths to the common ancestor of all putative cpFBPase1-type orthologs and all putative cpFBPase2-type orthologs is not markedly different. Therefore, the phylogenetic analyses cannot tell us which of the two cpFBPases originated first and was later duplicated to produce the other. The structural similarities between cyFBPase and cpFBPase2, however, suggest that cpFBPase 2 may have been the first chloroplastic FBPase. Specifically, both cyFBPase and cpFBPase2 lack the complete redox regulatory loop sequence and conserved Cys residues within the loop region. Thus, one likely scenario is that cyFBPase was first duplicated to produce cpFBPase2, which then was further duplicated to produce cpFBPase1. Subsequently, cpFBPase1 gradually acquired the redox-regulation domain during its evolutionary divergence.
The identification of cpFBPase2 in strawberry prompted researchers to speculate that cpFBPase2 may play crucial roles in specific tissues or processes such as development, stress response or metabolic regulation (Serrato et al., 2009). However, its subsequent discovery in the unicellular phytoflagellate E. gracilis (where it is called EgFBPaseII) clearly refutes the idea that cpFBPase2 plays roles in multicellular organismal function. More recently, research has shown that EgFBPaseII is expressed at extremely low levels and thus unlikely to contribute markedly to photosynthesis in Euglena cells under normal conditions. However, EgFBPaseII's greater tolerance to oxidative stress suggests that it is required under several oxidative stress conditions (Ogawa et al., 2015). Our findings here that cpFBPase2, similar to cpFBPase1, is ubiquitous in photosynthetic eukaryotes and is predicted to be localized in chloroplasts strongly imply that cpFBPase2 may also have a universal essential physiological function in chloroplasts.
We speculate that cpFBPase2 may participate in the Calvin cycle, similar to cpFBPase1, but under abnormal oxidative conditions caused by low light and low temperature. Evidence for this speculation is that FBPase can only catalyze the irreversible hydrolysis of fructose-1, 6-bisphosphate into fructose-6-phosphate and inorganic phosphate. Furthermore, cpFBPase2 acts independently of redox activation and is more tolerant of oxidation than cpFBPase1 (Ogawa et al., 2015; Serrato et al., 2009). Plants are often subject to oxidative stress at low temperatures under low light intensity (Haghjou et al., 2006), which is also a set of conditions that all photosynthetic eukaryotes often face. Although our speculation awaits further experimental verification, it is consistent with the conjecture of cpFBPase2's role in Euglena cells (Ogawa et al., 2015).
In conclusion, we found that cytosolic FBPase, cpFBPase 1, and cpFBPase2 are ubiquitously distributed in the major lineages of Archaeplastid and each form monophyletic groups, with cpFBPase1 and cpFBPase2 as sister groups. Taken together, these findings suggest that cpFBPase2 arose in the common ancestor of red and green algae, or even in the common ancestor of Archaeplastida or Plantae, and that all secondary plastid lineages inherited cpFBPase2 from their corresponding primary plastid lineage. We speculate that the ubiquitous coexistence of the two chloroplastic FBPases, cpFBPase1 and cpFBPase2, in photosynthetic eukaryotes is most likely the consequence of adaptive evolution to cope with different redox conditions during photosynthesis, especially those caused by recurrent changes in light conditions.
Conflict of interestThe authors declare that they have no competing interests.
Funding acknowledgementThis research was funded by the National Natural Science Foundation of China (31572256 and 31801967) and State Key Laboratory of Genetic Resources and Evolution (GREKF17-03).
Appendix A. Supplementary dataSupplementary data to this article can be found online at https://doi.org/10.1016/j.pld.2019.09.002.
Altschul S.F., Gish W., Miller W., Myers E.W., Lipman D.J., 1990. Basic local alignment search tool. J.Mol.Biol, 215: 403-410. DOI:10.1016/s0022-2836(05)80360-2 |
Armougom F., Moretti S., Poirot O., Audic S., Dumas P., Schaeli B., Keduas V., Notredame C., 2006. Expresso: automatic incorporation of structural information in multiple sequence alignments using 3D-coffee. Nucleic Acids Res, 34: W604-W608. DOI:10.1093/nar/gkl092 |
Blanc G., Duncan G., Agarkova I., Borodovsky M., Gurnon J., Kuo A., Lindquist E., Lucas S., Pangilinan J., Polle J., Salamov A., Terry A., Yamada T., Dunigan D.D., Grigoriev I.V., Claverie J.-M., Van Etten J.L., 2010. The Chlorella variabilis NC64A genome reveals adaptation to photosymbiosis, coevolution with viruses, and cryptic sex. Plant Cell, 22: 2943-2955. DOI:10.1105/tpc.110.076406 |
Bogen C., Al-Dilaimi A., Albersmeier A., Wichmann J., Grundmann M., Rupp O., Lauersen K.J., Blifernez-Klassen O., Kalinowski J., Goesmann A., Mussgnug J.H., Kruse O., 2013. Reconstruction of the lipid metabolism for the microalga Monoraphidium neglectum from its genome sequence reveals characteristics suitable for biofuel production. BMC Genomics, 14. DOI:10.1186/1471-2164-14-926 |
Brawley S.H., Blouin N.A., Ficko-Blean E., Wheeler G.L., Lohr M., Goodson H.V., Jenkins J.W., Blaby-Haas C.E., Helliwell K.E., Chan C.X., Marriage T.N., Bhattacharya D., Klein A.S., Badis Y., Brodie J., Cao Y., Collen J., Dittami S.M., Gachon C.M.M., Green B.R., Karpowicz S.J., Kim J.W., Kudahl U.J., Lin S., Michel G., Mittag M., Olson B.J.S.C., Pangilinan J.L., Peng Y., Qiu H., Shu S., Singer J.T., Smith A.G., Sprecher B.N., Wagner V., Wang W., Wang Z.-Y., Yan J., Yarish C., Zauner-Riek S., Zhuang Y., Zou Y., Lindquist E.A., Grimwood J., Barry K.W., Rokhsar D.S., Schmutz J., Stiller J.W., Grossman A.R., Prochnik S.E., 2017. Insights into the red algae and eukaryotic evolution from the genome of Porphyra umbilicalis (Bangiophyceae, Rhodophyta). Proc.Natl.Acad.Sci.U.S.A, 114: E6361-E6370. DOI:10.1073/pnas.1703088114 |
Brown G., Singer A., Lunin V.V., Proudfoot M., Skarina T., Flick R., Kochinyan S., Sanishvili R., Joachimiak A., Edwards A.M., 2009. Structural and biochemical characterization of the type II fructose-1, 6-bisphosphatase GlpX from Escherichia coli. J.Biol.Chem, 284: 3784-3792. DOI:10.1074/jbc.M808186200 |
Buchanan B.B., 1980. Role of light in the regulation of chloroplast enzymes. Annu.Rev.Plant Physiol.Plant Mol.Biol, 31: 341-374. DOI:10.1146/annurev.pp.31.060180.002013 |
Chiadmi M., Navaza A., Miginiac-Maslow M., Jacquot J.P., Cherfils J., 1999. Redox signalling in the chloroplast: structure of oxidized pea fructose-1, 6-bisphosphate phosphatase. EMBO J, 18: 6809-6815. DOI:10.1093/emboj/18.23.6809 |
Chueca A., Sahrawy M., Pagano E.A., Gorge J.L., 2002. Chloroplast fructose-1, 6-bisphosphatase: structure and function. Photosynth.Res, 74: 235-249. DOI:10.1023/a:1021243110495 |
Collen J., Porcel B., Carre W., Ball S.G., Chaparro C., Tonon T., Barbeyron T., Michel G., Noel B., Valentin K., Elias M., Artiguenave F., Arun A., Aury J.-M., Barbosa-Neto J.F., Bothwell J.H., Bouget F.-Y., Brillet L., Cabello-Hurtado F., Capella-Gutierrez S., Charrier B., Cladiere L., Cock J.M., Coelho S.M., Colleoni C., Czjzek M., Da Silva C., Delage L., Denoeud F., Deschamps P., Dittami S.M., Gabaldon T., Gachon C.M.M., Groisillier A., Herve C., Jabbari K., Katinka M., Kloareg B., Kowalczyk N., Labadie K., Leblanc C., Lopez P.J., McLachlan D.H., Meslet-Cladiere L., Moustafa A., Nehr Z., Collen P.N., Panaud O., Partensky F., Poulain J., Rensing S.A., Rousvoal S., Samson G., Symeonidi A., Weissenbach J., Zambounis A., Wincker P., Boyen C., 2013. Genome structure and metabolic features in the red seaweed Chondrus crispus shed light on evolution of the Archaeplastida.. Proc.Natl.Acad.Sci.U.S.A, 110: 5247-5252. DOI:10.1073/pnas.1221259110 |
Cseke C., Buchanan B.B., 1986. Regulation of the formation and utilization of photosynthate in leaves. Biochim.Biophys.Acta, 853: 43-63. DOI:10.1016/0304-4173(86)90004-2 |
Darriba D., Taboada G.L., Doallo R., Posada D., 2011. ProtTest 3: fast selection of best-fit models of protein evolution. Bioinformatics, 27: 1164-1165. DOI:10.1093/bioinformatics/btr088 |
Di Tommaso P., Moretti S., Xenarios I., Orobitg M., Montanyola A., Chang J.-M., Taly J.-F., Notredame C., 2011. T-Coffee: a web server for the multiple sequence alignment of protein and RNA sequences using structural information and homology extension. Nucleic Acids Res, 39: W13-W17. DOI:10.1093/nar/gkr245 |
Donahue J.L., Bownas J.L., Niehaus W.G., Larson T.J., 2000. Purification and characterization of glpX-encoded fructose 1, 6-bisphosphatase, a new enzyme of the glycerol 3-phosphate regulon of Escherichia coli. J.Bacteriol, 182: 5624-5627. DOI:10.1128/JB.182.19.5624-5627.2000 |
Emanuelsson O., Brunak S., von Heijne G., Nielsen H., 2007. Locating proteins in the cell using TargetP, SignalP and related tools. Nat.Protoc, 2: 953-971. DOI:10.1038/nprot.2007.131 |
Foflonker F., Price D.C., Qiu H., Palenik B., Wang S., Bhattacharya D., 2015. Genome of the halotolerant green alga Picochlorum sp reveals strategies for thriving under fluctuating environmental conditions. Environ.Microbiol, 17: 412-426. DOI:10.1111/1462-2920.12541 |
Gao C., Wang Y., Shen Y., Yan D., He X., Dai J., Wu Q., 2014. Oil accumulation mechanisms of the oleaginous microalga Chlorella protothecoides revealed through its genome, transcriptomes, and proteomes. BMC Genomics, 15. DOI:10.1186/1471-2164-15-582 |
Haghjou M.M., Shariati M., Pozveh M.H., 2006. The effect of low light intensities on oxidative stress induced by short-term chilling in Dunaliella salina teod. Pak.J.Biol.Sci, 9: 2048-2054. DOI:10.3923/pjbs.2006.2048.2054 |
Martin W., Mustafa A.Z., Henze K., Schnarrenberger C., 1996. Higher-plant chloroplast and cytosolic fructose-1, 6-bisphosphatase isoenzymes: origins via duplication rather than prokaryote-eukaryote divergence. Plant Mol.Biol, 32: 485-491. DOI:10.1007/bf00019100 |
Merchant S.S., Prochnik S.E., Vallon O., Harris E.H., Karpowicz S.J., Witman G.B., Terry A., Salamov A., Fritz-Laylin L.K., Maréchal-Drouard L., 2007. The Chlamydomonas genome reveals the evolution of key animal and plant functions.. Science, 318: 245-250. DOI:10.1126/science.1143609 |
Notredame C., Higgins D.G., Heringa J., 2000. T-Coffee: a novel method for fast and accurate multiple sequence alignment. J.Mol.Biol, 302: 205-217. DOI:10.1006/jmbi.2000.4042 |
Ogawa T., Kimura A., Sakuyama H., Tamoi M., Ishikawa T., Shigeoka S., 2015. Characterization and physiological role of two types of chloroplastic fructose-1, 6-bisphosphatases in Euglena gracilis. Arch.Biochem.Biophys, 575: 61-68. DOI:10.1016/j.abb.2015.04.002 |
Polle J.E.W., Barry K., Cushman J., Schmutz J., Tran D., Hathwaik L.T., Yim W.C., Jenkins J., McKie-Krisberg Z., Prochnik S., Lindquist E., Dockter R.B., Adam C., Molina H., Bunkenborg J., Jin E., Buchheim M., Magnuson J., 2017. Draft nuclear genome sequence of the halophilic and beta-carotene-accumulating green alga Dunaliella salina strain CCAP19/18. Genome Announc, 5. DOI:10.1128/genomeA.01105-17 |
Rashid N., Imanaka H., Kanai T., Fukui T., Atomi H., Imanaka T., 2002. A novel candidate for the true fructose-1, 6-bisphosphatase in archaea. J.Biol.Chem, 277: 30649-30655. DOI:10.1074/jbc.M202868200 |
Ronquist F., Teslenko M., van der Mark P., Ayres D.L., Darling A., Hohna S., Larget B., Liu L., Suchard M.A., Huelsenbeck J.P., 2012. MrBayes 3., 2: efficient bayesian phylogenetic inference and model choice across a large model space.. Syst.Biol., 61: 539-542. DOI:10.1093/sysbio/sys029 |
Schoenknecht G., Chen W.-H., Ternes C.M., Barbier G.G., Shrestha R.P., Stanke M., Braeutigam A., Baker B.J., Banfield J.F., Garavito R.M., Carr K., Wilkerson C., Rensing S.A., Gagneul D., Dickenson N.E., Oesterhelt C., Lercher M.J., Weber A.P.M., 2013. Gene transfer from bacteria and archaea facilitated evolution of an extremophilic eukaryote. Science, 339: 1207-1210. DOI:10.1126/science.1231707 |
Serrato A., Maria Yubero-Serrano E., Maria Sandalio L., Munoz-Blanco J., Chueca A., Luis Caballero J., Sahrawy M., 2009. cpFBPaseII, a novel redoxindependent chloroplastic isoform of fructose-1, 6-bisphosphatase. Plant Cell Environ, 32: 811-827. DOI:10.1111/j.1365-3040.2009.01960.x |
Stamatakis A., 2006. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics, 22: 2688-2690. DOI:10.1093/bioinformatics/btl446 |
Teich R., Zauner S., Baurain D., Brinkmann H., Petersen J., 2007. Origin and distribution of Calvin cycle fructose and sedoheptulose bisphosphatases in plantae and complex algae: a single secondary origin of complex red plastids and subsequent propagation via tertiary endosymbioses. Protist, 158: 263-276. DOI:10.1016/j.protis.2006.12.004 |
Verhees C.H., Kengen S.W.M., Tuininga J.E., Schut G.J., Adams M.W.W., De Vos W.M., Van der Oost J., 2003. The unique features of glycolytic pathways in Archaea. Biochem.J, 375: 231-246. DOI:10.1042/bj20021472 |
Ye Q., Tian H., Chen B., Shao J., Qin Y., Wen J., 2017. Giardia's primitive GPL biosynthesis pathways with parasitic adaptation 'patches': implications for Giardia's evolutionary history and for finding targets against Giardiasis. Sci.Rep, 7. DOI:10.1038/s41598-017-10054-1 |
Zimmermann G., Kelly G.J., Latzko E., 1978. Purification and properties of spinach leaf cytoplasmic fructose-1, 6-bisphosphatase. J.Biol.Chem, 253: 5952-5956. |



