Orchidaceae is one of the largest angiosperm families, consisting of more than 800 genera and about 25, 000 species (Dressler, 1993; Ramirez et al., 2007). Orchids are well known for their tremendous diversity in plant forms and growth habits, as well as extraordinary diversity in flower morphology and high value of hybrid cultivars (Ehlers et al., 2002; Givnish et al., 2015). Orchid production has therefore become one of the most important world-wide businesses in the floriculture industry. In China, Japan, Korea, and Southeast Asia, orchids in the genus Cymbidium are economically valued for their beautiful, fragrant flowers and elegant leaves. Cymbidium ensifolium, which belongs to the subgenus Jensoa, blossoms many times a year, and is thus highly desirable in China's flower market (Su et al., 2018a; Zhang et al., 2015).
Orchids are characterized by their highly evolved flower organs, which possess three petal-like sepals in the first whorl, with one in the top or dorsal sepal, and two lower lateral sepals. In the second whorl, there are two lateral petals and a specialized bottom petal, called a lip or labellum, which is oval to triangle and often has a kinked to undulated margin. In the middle of the flower there is a pistil/stigma fused together with pollinia to form the so-called column or gynostemium. Mutations that alter flower morphology are common in the orchid family and have greatly diversified orchid flower forms (Xiang et al., 2018; Yang et al., 2017). For example, in the multi-tepal mutants, the gynostemium is replaced by a newly emerged flower, which continues to produce sepals, petals, and gynostemium centripetally (Yang et al., 2017). In the stamenoidtepal mutant, outer tepals become shorter compared with those on the standard flower (Su et al., 2018b), and in the nongynostemium mutant, the gynostemium is absent and replaced by new sepals in the center part (Yang et al., 2017). Mutants with highly specialized floral morphology are not only valuable, but also represent valuable research materials for studying the molecular mechanisms of orchid flower pattern variation. However, the large genomes and long juvenile phases of orchids have greatly limited the utility of functional genomics and gene discovery approaches that might identify genes involved in the regulating floral patterning (Cai et al., 2015; Chao et al., 2017; Yan et al., 2015; Zhang et al., 2017).
| AP1 | APETALA1 |
| AP3 | APETALA3 |
| AGL | AGAMOUS-Like |
| bHLH | basic helix-loop-helix |
| bZIP | Basic leucine zipper |
| CO | CONSTANS |
| DEGs | Differentially expressed genes |
| GO | Gene ontology |
| KEGG | Kyoto encyclopedia of genes and genomes |
| MYB | Myeloblastosis transcription factors |
| NAC | NAM, ATAF, and CUC transcription factor |
| SPL | SQUAMOSA promoter binding protein-like |
| TALE | Three amino acid loop extension family |
| TCP | TEOSINTE BRANCHED1/CYCLOIDEA/PC transcription factors |
In this work, we examined a domesticated variety of C. ensifolium 'CuiYuMuDan', which we have named the "leaf-like mutant". This variety lacks typical characteristics of orchid floral organs such as zygomorphic flowers, the evolved lip, and the column; instead, these mutants produce sepal-to leaf-like structures along the inflorescence. By comparative analysis of the floral transcriptome between the wild type and the mutant, we identified 6234 differentially expressed genes (DEG) involved. GO and KEGG enrichment indicated that the unigenes responding to membranebuilding and anabolism regulation account for the majority. When referred to individual gene functions, we found 20 DEGs involved in flower development, including 13 MADS-box gene homologs, 3 CONSTANS (CO)-like genes, and other 4 unigenes that annotated as positive or negative regulator of floral meristem activity. Among them, floral homeotic MADS-box genes were investigated by phylogenetic analysis and expression validation. This study shed light on the molecular regulation of orchid flower development, particularly for floral organ defects, and provides new insights and practical guidance for improving their floral patterning regulation and molecular breeding.
2. Materials and methods 2.1. Plant materials and growth conditionsC. ensifolium wild-type plants (also known as 'XiaoTaoHong', the most widely known commercial cultivar in China) and the domesticated variety 'CuiYuMuDan', were artificially cultivated and collected from the cultivation base of the Environmental Horticulture Research Institute, Guangdong Academy of Agricultural Sciences, China. All plants were grown and maintained in pots in a greenhouse at day/night temperatures of 26/23 ℃ under a 16-h light/8-h dark photoperiod.
2.2. Library construction and illumina sequencingThe cDNA libraries of wild-type C. ensifolium 'XiaoTaoHong' and the mutant plant 'CuiYuMuDan' were prepared from an equal mixture of RNAs isolated from mature flowers of five individual plants, respectively. Two replications were included in each sample to create four multiplexed cDNA libraries. Briefly, mRNAs were purified from total RNA using the Oligotex mRNA Midi Kit (QIAGEN, Germany) and quantified using a Nano-Drop 2000 spectrophotometer (Thermo Scientific, USA) to generate the cDNA library according to the Illumina manufacturer's instructions as in our previous work (Yang and Zhu, 2015). MRNAs were purified from total RNA and fragmented to approximately 200 bp. Subsequently, the collected mRNAs were subjected to first strand and second strand cDNA synthesis followed by adaptor ligation and enrichment with a low-cycle according to the instructions of the TruSeq® RNA HT Sample Prep Kit (Illumina, USA). The purified library products were evaluated using the Agilent 2200 TapeStation and Qubit®2.0 (Life Technologies, USA) and then diluted to 10 pM for cluster generation in situ on the HiSeq3000 pair-end flow cell followed by sequencing (2 × 100 bp). An average of more than 75 million reads was generated for each sample.
2.3. Analysis of differentially expressed genesGene expression levels were calculated by FPKM values: FPKM =[total transcript fragments/mapped fragments (millions)] × transcript length (kb). Significant differences in gene expression between wild type and mutant were determined using edgeR. The false discovery rate (FDR) was applied to identify the threshold of the P-value in multiple tests; with FDR × 0.05 and|log2 ratio|> 1 (two-fold change) as the threshold for a significant difference in gene expression.
Differentially expressed genes (DEGs) were annotated using gene ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analyses following Yang and Zhu (2015) and Yang et al. (2017). First, all DEGs were mapped to GO terms (or KEGG pathways) in the databases (http://www.geneontology.org/ or http://www.genome.ad.jp/) and gene numbers were calculated for every term (or pathway) (Conesa et al., 2005; Kanehisa and Goto, 2000). This was followed by a hypergeometric test to find the significantly enriched terms in DEGs compared to the genome background. For this purpose, we used the corrected P-value ≤ 0.05 as a threshold for the significantly enriched GO terms and Q-value ≤ 0.05 as a threshold for KEGG pathways.
2.4. Real-time quantitative RT-PCR (qRT-PCR) analysisOne μg of total RNA was quantified with a NanoDrop 2000 spectrophotometer (Thermo Scientific) and used for reverse-transcription following an oligo(dT) primed cDNA synthesis protocol (Fermentas). The resulting cDNA was subjected to relative quantitative PCR using Bio-Rad CFX-96 RealTime PCR System (Bio-Rad, USA) in a final volume of 20 μl containing 2 μl of cDNA and 10 μl of SYBR premix Ex-taqTM (Takara, Japan). To compare gene expression levels between accessions, ubiquitin was used as an internal control for normalization. A relative quantitative method (DDCt) was used to evaluate the quantitative variation. For each reported result at least three independent biological samples were subjected to a minimum of three technical replicates. The primers were designed with Primer 5.0 software and are listed in Table S1.
2.5. Multiple sequence alignment and phylogenetic analysisA total of 108 complete protein sequences were aligned using MUSCULE, including 7 CeMADS genes from C. ensifolium, 26 AsMADS from Apostasia, 28 DcMADS from Dendrobium catenatum, 25 EpMADS from Erycina pusila and 22 PEQUMADS from Phalaenopsis equestris. Based on the alignment, a Maximum likelihood (ML) phylogenetic tree was constructed using MEGAX with 1000 bootstrap value and Jones-Taylor-Thornton method (Kumar et al., 2016). Genbank accession number of MADS-box genes used in phylogenetic analysis are listed in Table S2.
2.6. Scanning electron microscopyDissected apices of mature flowers were fixed in a solution of 3% glutaraldehyde and 2% formaldehyde for 24 h. Samples were then dehydrated in acetone, critical-point dried in liquid CO2, and mounted on stubs and sputter coated with 25 nm gold. Samples were examined using a JSM-6360LV (JEOL) scanning electron microscope.
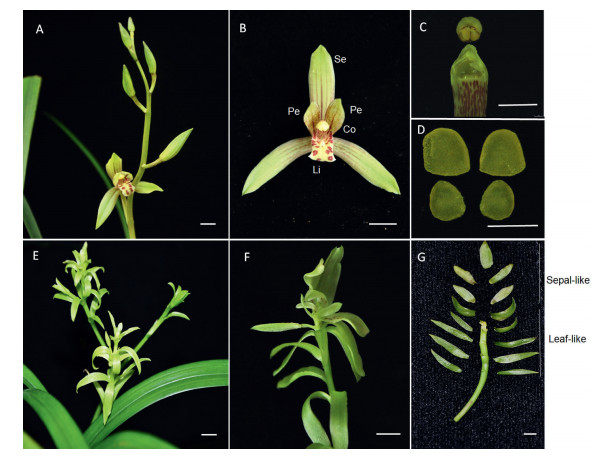
3. Results 3.1. Morphology of Cymbidium ensifolium leaf-like flower mutantThe wild-type flowers of C. ensifolium 'XiaoTaoHong' have three sepals in the first whorl and three petals in the second whorl. Together these are called the tepals. Two of the lateral petals have the same shape, and the bottom petal which is called the labellum (or lip) is highly evolved, with an ovate to triangular shape (Fig. 1A and B). The male and female reproductive organs are highly fused to form a gynostemium, which evolved through the complete fusion of the style, stigma and staminal filament, and has four pollinia on a semi-circular viscidium (Fig. 1C and D). In comparison, these typical characteristics of orchid flowers are absent from the mutant plant 'CuiYuMuDan' and differentiation of floral organs is severely inhibited. Instead of differentiated floral organs, the leaf-like flower mutant has sepal-to leaf-like structures along the inflorescence (Fig. 1E-G). To identify differences in the shapes of wild type and mutant floral organs, we observed perianth epidermal cells with a scanning electron microscope. In wild-type flowers, the epidermal cell shapes and ultrastructural features of wild-type sepals, petals, lips, and columns vary (Fig. 2A-H). On both the adaxial and abaxial epidermis, sepal cells are polygonal; petal cells are hemispherical; lip cells are spherical; and column cells are papillae/cupola/conica. In contrast, mutant sepal and petal-like structures (Fig. 1, G) have no polygonal to ellipticalshaped cells; instead, consistent with the abnormal smooth surface of the leaf-like flower structures, mutant flowers have flat epidermal cells (Fig. 2, I & L).
|
| Fig. 1 Flower morphology of wild type (A-D) and mutant (E-G). Se, sepal primordium; Pe, petal primordium; Li, lip primordium; Co, column primordium. Bar = 1 cm. |

|
| Fig. 2 Scanning electron micrographs of perianth epidermal cells of wild type and mutant plants. (A-D): Adaxial epidermis in the wild type flower sepal, petal, lip and column. (E-H): Abaxial epidermis in the wild type flower sepal, petal, lip, and the column. (I-L): Perianth epidermal cells of the mutant: I & J: Adaxial epidermis in the leaf- and sepal-like structures. K & L: Abaxial epidermis in the leaf- and sepal-like structures. Scale bars: 100 μm. |
Transcriptome sequencing generated 77, 520, 950 total reads for the wild-type 'XiaoTaoHong' (WT_Xth) and 85, 119, 788 for the leaf-like flower mutant 'CuiYuMuDan' (Mut_Cymd), with corresponding clean reads of 76, 749, 454 (99.00%) and 84, 486, 426 (99.26%) (Table 1). Currently, no C. ensifolium reference genome sequence exists; thus, we assembled all clean reads (25, 748, 328, 939 bp) de novo and obtained a dataset of 44, 480 unigenes, which had a mean length of 891 bp (NCBI Genbank accession number SRR9117944, SRR9117943 and Table S1). This data set is smaller than the available ESTs derived from C. ensifolium flowers in our previous report (Yang and Zhu, 2015). We therefore mapped all sequencing reads to 111, 892 unigenes reported in our previous work. The total mapped reads for WT_Xth and Mut_Cymd were 68, 700, 479 (89.51%) and 60, 198, 127 (80.08%), respectively. This suggests that most transcriptionally active genes were captured in our initial transcriptome data set.
| WT_Xth | Mut_Cymd | |
| Total reads | 77, 520, 950 | 85, 119, 788 |
| Clean reads | 76, 749, 454 (99.00%) | 84, 486, 426 (99.26%) |
| Clean_reads_base (bp) | 12, 229, 919, 375 (97.27%) | 13, 518, 409, 564(97.98%) |
| Total mapped | 68, 700, 479 (89.51%) | 60, 198, 127 (80.08%) |
| Multiple mapped | 13, 931, 494 (18.15%) | 22, 558, 363 (30.01%) |
| Uniquely mapped | 54, 768, 985 (71.36%) | 37, 639, 764 (50.07%) |
To identify the transcriptional responses in the mutant flower, we determined FPKM values for each gene and calculated the number of DEGs (FPKM ≥1). Overall, 6234 transcripts showed significantly altered expression in the mutant, with 3578 up-and 2656 down-regulated (Table S3). These findings suggest that these DEGs may play a role in leaf-like flower development.
Numbers inside the parentheses indicate the percentage of total reads. Total mapped:Number of reads mapped to the reference dataset; Multiple mapped:Number of reads with multiple alignment sites; Uniquely mapped:Number of reads with single alignment site.
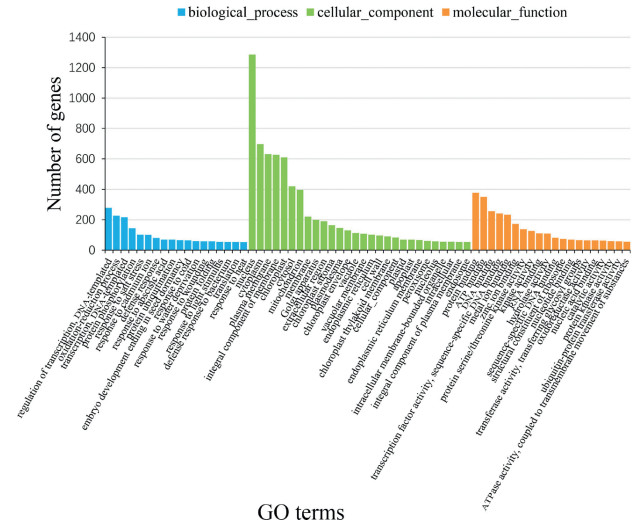
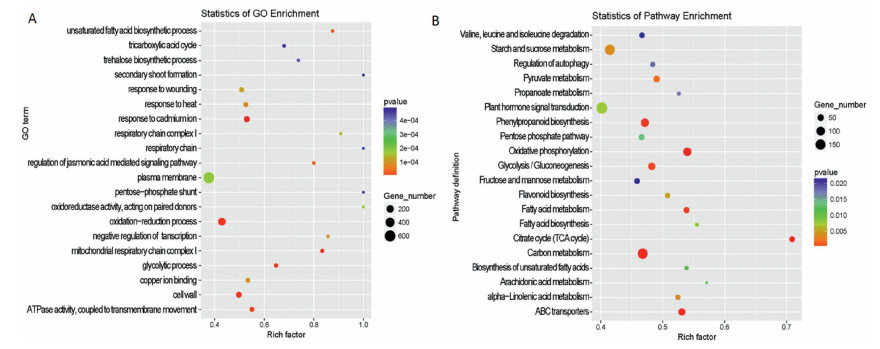
3.3. Annotation statistics and functional enrichmentGO enrichment analysis revealed that leaf-like flower mutant DEGs can be categorized into 66 functional groups. Of these, 18 GO terms were related to 'biological process', 28 were related to 'cellular component', and 20 were related to 'molecular functions. Within each of the three main categories of the GO classification scheme, the dominant subcategories were 'regulation of transcription & DNA-templated' (278), 'nucleus' (1286) and 'protein binding' (377), respectively. In addition, we noticed that a relatively high-percentage of genes came from the GO terms of 'cytoplasm', 'plasma membrane', 'integral component of membrane' and 'chloroplast' in the 'cellular component' categories (Fig. 3). After normalization to the total number of assigned genes for each of the 66 GO terms, we found significant enrichment occurred in 'secondary shoot formation', 'respiratory chain', 'pentose-phosphate shunt', and 'oxidoreductase activity' (Fig. 4A).
|
| Fig. 3 GO classification of unigenes differentially expressed between wild type and mutant. |
|
| Fig. 4 The scatter plot of GO clusters (A) and KEGG pathways (B) significantly enriched in the mutant. |
We also conducted an enrichment analysis using KEGG path-ways, which assigned 4239 DEGs to 134 pathways. Of these, the majority of hits fell into 'plant-pathogen interaction (184)' and 'plant hormone signal transduction' (182), followed by 'starch and sucrose metabolism' (149) and 'carbon metabolism' (146) (Table S4). However, the most significant enrichment was detected in the 'citrate cycle' (Fig. 4, B). These findings indicate that the 'citrate cycle' pathway, which functions as the ultimate metabolic pathway of the three major nutrients (carbohydrates, lipids and amino acids), is involved in the regulation of leaf-like flower development. More details of the pathway annotations for significant hits in unigene sets are provided in Table S4.
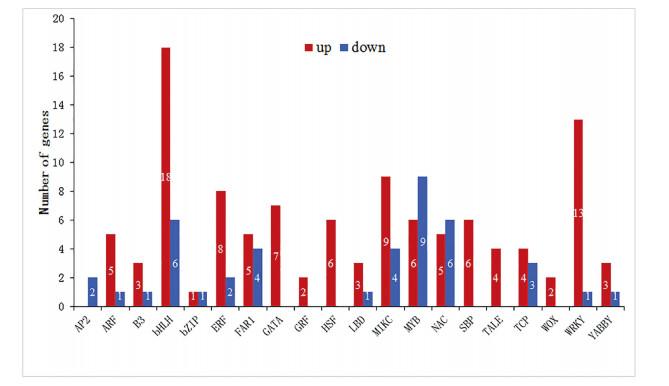
3.4. Differential expression of transcription factorsTranscription factors play a critical role in the control of plant reproductive development. In our study, a total of 152 putative transcription factors were detected with significant changes in expression between wild-type and the mutant, with 110 up-and 42 down-regulated, respectively (Table S5). We found that the genes that showed the greatest differences in expression were transcription factors in the bHLH family (24), followed by MYB (15), WRKY (14), and MIKC families (13) suggesting that transcription factors in these gene families play a crucial role in C. ensifolium flower development (Fig. 5). We also noted expression differences in NAC, TCP, SBP and TALE transcription factor families. Expression differences in these developmentally important transcription factor families suggest that genes in these families may play roles in leaf-like flower mutant defects in floral organ development.
|
| Fig. 5 Transcription factors differentially expressed between wild type and mutant. Number of up-regulated (red) and down-regulated (blue) transcripts were quantified in the mutant compared to the wild type (based on the FPMK value with |log2 (fold change)| > 1 and Q-value < 0.05). |
We specifically searched for the orthologs of genes and gene families that are known to be involved in flower initiation and the ABCDE flower developmental model of Arabidopsis. According to the unigene annotations, we identified 20 genes involved in flower development. Eleven of these gene were up-regulated, whereas nine genes were down regulated. These DEGs associated with floral development included 13 unigenes which were identified as MADS-box gene homologs, 3 CONSTANS (CO)-like genes, and other four unigenes that were annotated as positive or negative regulators of floral meristem activity, including CLAVATA3 (CLV3), WUSCHEL (WUS) and PERIANTHIA (PAN) (Somssich et al., 2016; Wang et al., 2017; Uemura et al., 2018). Expression of two genes homologous to WUS increased by 1.49-and 1.09-fold respectively, whereas two unigenes with CLV3-like and PAN-like sequences, were expressed at lower levels in the mutant (Table 2). According to reports on Arabidopsis, WUS is required to maintain stem cells in an undifferentiated state. The size of the WUS expression domain controls the size of the stem cell population through WUS indirectly activating the expression of CLV3 in the stem cells and CLV3 repressing WUS transcription through the CLV1 receptor kinase signaling pathway (Guo et al., 2018; Uemura et al., 2018). Thus, our findings that, in the mutant, WUS-like sequences are expressed at high levels and CLV3- like genes are expressed at low levels is consistent with sustained differentiation of the inflorescence axis.
| Trans_ID | Annotation | Mut | WT_Xth | Mut_ FPKM | WT_Xth _FPKM | log2FC | Regulation | P value |
| MADS-box genes | ||||||||
| DN29274_c0_g4 | SEP-like MADS-box protein [Cymbidium ensifolium] |
302.00 | 13191.00 | 6.09 | 290.2 | -5.83 | down | 0.00 |
| DN28990_c1_g1 | class E MADS-domain transcription factor, partial [Gongora galeata] |
1191.03 | 6581.26 | 30.23 | 171.79 | -2.84 | down | 0.00 |
| DN23421_c0_g1 | DEFICIENS-like MADS-box transcription factor [Gongora galeata] |
1867.00 | 6600 | 243.09 | 1029 | -2.20 | down | 0.00 |
| DN23421_c0_g2 | MADS5 transcription factor [Phalaenopsis equestris] | 1299.94 | 3197.95 | 38.22 | 100.02 | -1.68 | down | 0.00 |
| DN27680_c1_g5 | MADS-box protein 12 [Erycina pusilla] | 420.00 | 153.00 | 25.57 | 13.07 | 1.08 | up | 0.00 |
| DN32011_c0_g2 | PREDICTED: agamous-like MADS-box protein AGL61 [Camelina sativa] |
830.00 | 43.00 | 60.12 | 4.1 | 3.89 | up | 0.00 |
| DN23236_c1_g3 | MADS40 [Erycina pusilla] | 534.72 | 72.00 | 14.34 | 2.37 | 2.51 | up | 0.00 |
| DN28371_c0_g1 | MADS box protein, partial [Rhynchostylis retusa] | 408.07 | 71.85 | 77.83 | 16.72 | 2.12 | up | 0.00 |
| DN23985_c2_g1 | MADS box protein, partial [Rhynchostylis retusa] | 329.00 | 79.00 | 100.62 | 24.8 | 1.68 | up | 0.00 |
| DN27504_c0_g4 | MADS box protein, partial [Rhynchostylis retusa] | 217.50 | 34.81 | 114.77 | 20.07 | 2.26 | up | 0.00 |
| DN23829_c3_g2 | AP1-like protein [Cymbidium faberi] | 1133.00 | 92.00 | 21.73 | 1.90 | 3.24 | up | 0.00 |
| DN30954_c0_g1 | hypothetical MADS box protein Csa_6G076710 [Cucumis sativus] |
1468.99 | 290.00 | 246.69 | 52.76 | 1.96 | up | 0.00 |
| DN27680_c1_g5 | MADS-box protein 12 [Erycina pusilla] | 420.00 | 153.00 | 25.57 | 13.07 | 1.08 | up | 0.00 |
| Gene regulating flowering time | ||||||||
| DN30019_c0_g1 | CONSTANS-like 6 [Erycina pusilla] | 871.95 | 2209.89 | 19.22 | 53.18 | -1.72 | down | 0.00 |
| DN31348_c1_g2 | CONSTANS-like 8 [Erycina pusilla] | 2771.00 | 5955.00 | 61.95 | 147 | -1.48 | down | 0.00 |
| DN21959_c0_g1 | CONSTANS-like 10 [Erycina pusilla] | 1795.00 | 2895.00 | 29.41 | 70.24 | -1.07 | down | 0.00 |
| Positive and genative regulator of floral meristem activity | ||||||||
| DN29284_c1_g5 | CLAVATA3/ESR (CLE)-related protein 25-like [Phoenix dactylifera] |
196.38 | 317.55 | 5.21 | 8.82 | -1.08 | down | 0.00 |
| DN29072_c2_g2 | PREDICTED: WUSCHEL-related homeobox 8-like [Elaeis guineensis] |
3264.00 | 893.00 | 66.34 | 19.97 | 1.49 | up | 0.00 |
| DN29072_c2_g4 | PREDICTED: WUSCHEL-related homeobox 8-like [Elaeis guineensis] |
141.00 | 51.00 | 3.72 | 1.47 | 1.09 | up | 0.00 |
| DN26944_c0_g2 | PREDICTED: transcription factor TGA2.3-like [Musa acuminata] |
1123.00 | 1892.00 | 14.95 | 27.14 | -1.13 | down | 0.00 |
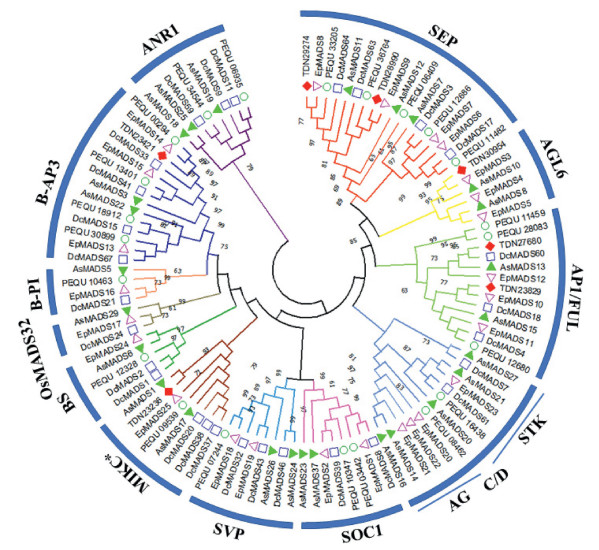
MADS-box genes are known to play key roles in flower development; we therefore analyzed MADS-box genes that were differentially expressed in the leaf-like flower mutant. We used 50RACE to sequence complete MADS-box genes (Table S6) and then constructed a maximum likelihood (ML) phylogenetic tree of 108 protein sequences from four orchid species. Seven Cymbidium ensifolium MADS-box genes were clustered with floral homeotic genes isolated from other orchid species:two AP1/FUL homologs, two SEP homologs, and one putative ortholog each of AGL6, AP3 and MIKC* genes (Fig. 6).These gene sequences could be used as an important resource for future studies on floral development and flower organ formation.
|
| Fig. 6 Phylogenetic analysis of the MADS-box transcription factors from different orchid plant species. Amino acid sequences were aligned by MUSCULE, and phylogenetic relationships were reconstructed using a maximum-likelihood (ML) method in MEGAX software with JTT amino acid substitution model. Bootstrap values for 1000 replicates were used to assess the robustness of the trees. Previously published MADS-box protein sequences of Phalaenopsis equestris, Dendrobium catenatum, Erycina pusila and Apostasia shenzhenica were retrieved from GenBank and the accession number listed in Table S2. |
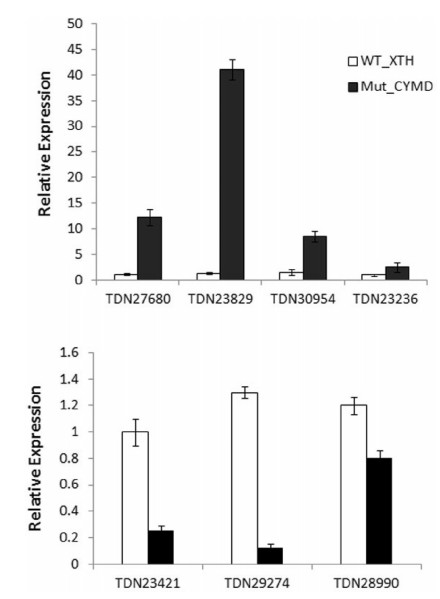
We used real-time quantitative RT-PCR to validate the differential expression of these seven MADS-box genes. A-class AP1 homologous genes TDN27680 and TDN23829 were expressed 12.3- and 41-fold higher, respectively, in the mutant than in the wildtype. Similarly, AGL6-like TDN30954 and MIKC* gene TDN23236 were expressed 8.5-and 2.5-fold higher, respectively, in the mutant than that in the wild-type flowers (Fig. 7). In contrast, B-class AP3 homolog TDN23421 and E-class SEP-like sequences TDN29274 and TDN28990 were expressed 0.9-to 0.3-fold less in the mutant than in the wild-type. The differential expression of these floral homeotic genes between mutant and wild-type flowers is consistent with our initial analysis of differential gene expression.
|
| Fig. 7 Quantitative RT-PCR analysis of MADS-box genes. The y-axis indicates fold change in expression among the samples. Expression levels were normalized using the threshold cycle values obtained for the Ubiquitin and Actin genes. Error bars indicate the standard deviation of the mean (SD) (n = 3). Three replicates were analyzed, with similar results. One-way ANOVA with Bonferroni multiple comparison tests significant at P < 0.05 between the two samples. |
Leaf-like flower variations occur frequently in angiosperms. The genes that are thought to regulate these variations (e.g., LEAFY, AGL24, SVP, phytoplasma effector SAP54) have been well documented in several plant species, including Arabidopsis, rice, and tomato (Ferrario et al., 2004; Fornara et al., 2008; Li et al., 2016; MacLean et al., 2011; Schultz and Haughn, 1991; Yu et al., 2004). Numerous economically valuable Cymbidium varieties have been produced that have diverse inflorescence architectures, including leaf or flower color and floral patterning. Nevertheless, few studies have examined the molecular mechanisms of floral organ development and variation in Cymbidium spp.
During the past decade, genomics approaches have been widely applied to horticultural crops, and several Cymbidium EST libraries have been reported (Sun et al., 2016; Yang and Zhu, 2015; Yang et al., 2017, 2019). Application of next-generation sequencing technologies has greatly advanced the study of orchid genomics. Recently we published a transcriptome data set containing 111, 892 C. ensifolium transcript clusters. In this study, we dissected the C. ensifolium leaf-like flower mutant for the first time. Comparative transcriptome analysis identified 6234 DEGs, most of which are involved in cell division and anabolism. Gene Ontology terms enriched for defective flowers were related to 'mitochondrion', 'plasma membrane', 'nucleus', and 'RNA transport'; whereas, KEGG pathways were related to 'Starch and sucrose metabolism', 'Carbon metabolism', 'Biosynthesis of amino acids', 'Protein processing in endoplasmic reticulum', 'Phenylpropanoid biosynthesis', 'RNA transport', and 'Amino sugar and nucleotide sugar metabolism'. The differential expression of genes involved in plant cell expansion and energy metabolism is consistent with the developmental processes that underlie flower development, i.e., cell division, membranebuilding, and regulation of anabolism.
We also identified transcription factors that correspond to floral organ defects in the mutant. Specifically, we identified 110 transcription factors that are up-regulated in the leaf-like flower mutant and 42 transcription factors that are down-regulated. The majority of these genes belong to the transcription factor family bHLH followed by MYB, WRKY, and MIKC gene families. The differential expression of these genes in mutant flowers suggest that they play crucial roles in C. ensifolium flower development. In addition, expression differences were detected in developmentally important transcription factor families such as NAC, TCP, SBP, and TALE. Taken together, these findings suggest that flower development is regulated by a complex regulatory network of genes. For example, the NAC genes CUC1 and CUC2 have been shown to be involved in petal initiation and petal number determination; furthermore, other NAC members may function in different stages of petal development, such as petal initiation and petal expansion (Cucinotta et al., 2018; Spinelli et al., 2011). TCP proteins have important functions in plant development, such as control of leaf shape and flower symmetry, and the suppression of lateral shoot branching (Nicolas and Cubas, 2016a, 2016b). In addition, TALE genes are required for meristem maintenance and proper patterning of organ initiation, and play a critical role in the diversity of flower and leaf form (Furumizu et al., 2015; Tao et al., 2018). The regulatory relationship between SPL proteins and the meristem identity AP1/FUL-like genes has also been well documented (Chen et al., 2018; Liu et al., 2017).
We further characterized differentially expressed genes from the MIKC type MADS-box transcription factor family, including two AP1/FUL homologs, two SEP homologs, one putative ortholog of AGL6, AP3, and MIKC*. Quantitative RT-PCR confirmed that in the mutant A-class AP1 homologous genes TDN27680 and TDN23829, AGL6-like TDN30954, and MIKC* gene TDN23236 were highly expressed, whereas expression of B-class AP3 homolog TDN23421 and E-class SEP1/3-clade genes TDN29274 and TDN28990 decreased.
Previous studies on the regulation of flower organ development have noted that perianth patterning in orchids is mediated by ABCE model-related MADS-box genes. For example, during flower patterning in Phalaenopsis, PeMADS6/PeMADS2-5 (PI-like/AP3- like) form important heterodimeric complexes; in Dendrobium DcOSEP1/DcOPI/DcOAP3A or DcOAP3B (SEP-like/PI-like/AP3-like) form multimeric proteins; similarly, in Oncidium, AGAMOUS-LIKE6- like (AGL6-like) and AP3-like proteins form multimeric proteins; and in Habenaria radiata, HrDEF-C3/HrAGL6-C2 form complexes (Tsai et al., 2004; Chang et al., 2010; Hsu et al., 2015; Mitoma et al., 2019). Notably, SEP genes serve as glue factors to interact with the mass of multi-MADS-box proteins and facilitate the formation of various protein complexes during sepal, petal, and lip determination. Pan et al. (2014) revealed that in Phalaenopsis dimeric, trimeric and tetrameric protein-protein interactions form between PeSEP and other MADS-box proteins, such that participation of these four SEP paralogs in Phalaenopsis varies during floral transition, morphogenesis, and ovule development. For example, PeSEP3 has a significant effect on floral morphology. Virus-induced silencing of PeSEP3 alters the epidermal identity of tepals and the contents of anthocyanin and chlorophyll, causing tepals to become leaf-like organs. In our work, the expression of CeSEP1/3-clade genes TDN29274 and TDN28990, the orthologs of PeSEP1 and PeSEP3, respectively, was reduced by 30-90%. However, the other two CeSEP genes showed equal or slightly higher expression in the mutant (data not shown). This result agrees well with research from Phalaenopsis that shows that defects of CeSEP1/3-clade genes contributes to the leaf-like flower phenotype in the mutant. Furthermore, these results lend support to the idea that SEP paralogs differ in their ability to regulate floral organ specificity.
This is the first study to identify effectors of leaf-like flower development. These transcriptome data sets provide a significant resource for the discovery of genes related to floral patterning and improve our understanding of the floral morphological diversity of the orchid family.
Declaration of Competing InterestWe would like to submit the enclosed manuscript entitled "Transcriptome Analysis Reveals Clues into Leaf-Like Flower Development in Chinese Orchid C. ensifolium", which we wish to be considered for publication in Plant Diversity. No conflict of interest exits in the submission of this manuscript, and the manuscript is approved by all authors for publication. I would like to declare on behalf of my co-authors that the work described was original research that has not been published previously, and not under consideration for publication elsewhere, in whole or in part.
In this work, we dissected a domesticated variety of C. ensifolium 'CuiYuMuDan', which continuous to produce sepal-to leaf-like structures and named as leaf-like flower mutant accordingly. Using comparative transcriptome analysis, we identified 6234 differentially expressed genes (DEGs) mainly involved in membranebuilding, anabolism regulation, and plant hormone signal transduction implying their potential roles involved in floral development deficiency in the mutant. When annotated with plant transcription factor database, we found 152 differentially expressed transcription factors and the most differences occurred in the families of bHLH, MYB, MIKC, and WRKY. Moreover, when referred to individual gene functions, we found 20 DEGs involved in flower development, including MADS-box genes, CLAVATA3 (CLV3), WUSCHEL (WUS) and PERIANTHIA (PAN). Among them, 7 floral homeotic genes were further investigated by phylogenetic analysis and expression validation, which displayed distinctive spatial expression patterns and significant changes between the wild type and the mutant. Our result shed light on the molecular regulation of orchid flower development, particularly represents the first report on transcriptome analysis of floral organ deficiency in C. ensifolium, and provides new insights and practical guidance for improving floral patterning regulation and molecular breeding of Chinese orchid plants. So we believe that our findings could be of interest to the readers of Plant Diversity, and we hope that the editorial board will agree on the interest of this study.
We deeply appreciate your consideration of our manuscript, and we look forward to receiving comments from the reviewers. If you have any queries, please don't hesitate to contact me at the address below.
AcknowledgementsThis research was funded by grants from National Key R & D Program (2018YFD1000404), the National Natural Science Foundation of China (31672184), the Natural Science Foundation of Guangdong Province (2017A030312004), Guangzhou Science and Technology Project (201707010307, 201904020026), Innovation Team of Modern Agricultural Industry Technology System in Guangdong Province (2019KJ121), and the Guangdong Academy of Agricultural Sciences Discipline Team Construction Project (201612TD, 2017A070702008, 201721).
Appendix A. Supplementary dataSupplementary data to this article can be found online at https://doi.org/10.1016/j.pld.2019.12.001.
Cai J., Liu X., Vanneste K., Proost S., Tsai W.C., Liu K.W., Chen L.J., He Y., Xu Q., Bian C., et al, 2015. The genome sequence of the orchid Phalaenopsis equestris. Nat.Genet, 47: 65-72. DOI:10.1038/ng.3149 |
Chang Y.Y., Kao N.H., Li J.Y., Hsu W.H., Liang Y.L., Wu J.W., et al, 2010. Characterization of the possible roles for B class MADS box genes in regulation of perianth formation in orchid. Plant Physiol, 152: 837-853. DOI:10.1104/pp.109.147116 |
Chao Y.T., Yen S.H., Yeh J.H., Chen W.C., Shih M.C., 2017. Orchidstra 2.0-A transcriptomics resource for the orchid family.. Plant Cell Physiol, 58: e9. |
Chen D., Yan W., Fu L.Y., Kaufmann K., 2018. Architecture of gene regulatory networks controlling flower development in Arabidopsis thaliana. Nat.Commun, 9. |
Conesa A., Gotz S., Garcia-Gomez J.M., Terol J., Talon M., Robles M., 2005. Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research.. Bioinformatics, 21: 3674-3676. DOI:10.1093/bioinformatics/bti610 |
Cucinotta M., Manrique S., Cuesta C., Benkova E., Novak O., Colombo L., 2018. CUP-SHAPED COTYLEDON1 (CUC1) and CUC2 regulate cytokinin homeostasis to determine ovule number in Arabidopsis. J.Exp.Bot, 69: 5169-5176. DOI:10.1093/jxb/ery281 |
Dressler, R., 1993. Phylogeny and Classification of Orchid Family. Cambridge University Press, Cambridge, pp. 102-218.
|
Ehlers B.K., Olesen J.M., Agren J., 2002. Floral morphology and reproductive success in the orchid Epipactis helleborine: regional and local across-habitat variation. Plant Systemat.Evol, 236: 19-32. DOI:10.1007/s00606-002-0197-x |
Ferrario S., Busscher J., Franken J., Gerats T., Vandenbussche M., Angenent G.C., Immink R.G., 2004. Ectopic expression of the petunia MADS box gene UNSHAVEN accelerates flowering and confers leaf-like characteristics to floral organs in a dominant-negative manner.. Plant Cell, 16: 1490-1505. DOI:10.1105/tpc.019679 |
Fornara F., Gregis V., Pelucchi N., Colombo L., Kater M., 2008. The rice StMADS11-like genes OsMADS22 and OsMADS47 cause floral reversions in Arabidopsis without complementing the svp and agl24 mutants. J.Exp.Bot, 59: 2181-2190. DOI:10.1093/jxb/ern083 |
Furumizu C., Alvarez J.P., Sakakibara K., Bowman J.L., 2015. Antagonistic roles for KNOX1 and KNOX2 genes in patterning the land plant body plan following an ancient gene duplication. PLoS Genet, 11: e1004980. DOI:10.1371/journal.pgen.1004980 |
Givnish T.J., Spalink D., Ames M., Lyon S.P., Hunter S.J., Zuluaga A., Iles W.J., Clements M.A., Arroyo M.T., Leebens-Mack J., et al, 2015. Orchid phylogenomics and multiple drivers of their extraordinary diversification.. P Roy Soc Lond B Bio, 1814: 282. |
Guo L., Cao X., Liu Y., Li J., Li Y., Li D., Zhang K., Gao C., Dong A., Liu X., 2018. A chromatin loop represses WUSCHEL expression in Arabidopsis. Plant J, 94: 1083-1097. DOI:10.1111/tpj.13921 |
Hsu H.F., Hsu W.H., Lee Y.I., Mao W.T., Yang C.H., 2015. Model for perianth formation in orchids. Native Plants, 1: 15046. DOI:10.1038/nplants.2015.46 |
Kanehisa M, Goto S, 2000. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res, 28: 27-30. DOI:10.1093/nar/28.1.27 |
Kumar S., Stecher G., Tamura K., 2016. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets.. Mol.Biol.Evol, 33: 1870-1874. DOI:10.1093/molbev/msw054 |
Li Z., Zeng S., Li Y., Li M., Souer E., 2016. Leaf-like sepals induced by ectopic expression of a SHORT VEGETATIVE PHASE (SVP)-Like MADS-box gene from the basal eudicot epimedium sagittatum. Front.Plant Sci, 7: 1461. |
Liu N., Tu L., Wang L., Hu H., Xu J., Zhang X., 2017. MicroRNA 157-targeted SPL genes regulate floral organ size and ovule production in cotton. BMC Plant Biol, 17: 7. DOI:10.1186/s12870-016-0969-z |
MacLean A.M., Sugio A., Makarova O.V., Findlay K.C., Grieve V.M., Toth R., Nicolaisen M., Hogenhout S.A., 2011. Phytoplasma effector SAP54 induces indeterminate leaf-like flower development in Arabidopsis plants. Plant Physiol, 157: 831-841. DOI:10.1104/pp.111.181586 |
Mitoma M., Kajino Y., Hayashi R., Endo M., Kubota S., Kanno A., 2019. Molecular mechanism underlying pseudopeloria in Habenaria radiata (orchidaceae). Plant J, 99: 439-451. DOI:10.1111/tpj.14334 |
Nicolas, M., Cubas, P., 2016a. The role of TCP transcription factors in shaping flower structure, leaf morphology, and plant architecture. In: Gonzalez, D.H. (Ed.), Plant Transcription Factors. Academic Press, Boston, pp. 249-267.
|
Nicolas M., Cubas P., 2016b. TCP factors: new kids on the signaling block. Curr.Opin.Plant Biol, 33: 33-41. DOI:10.1016/j.pbi.2016.05.006 |
Pan Z.J., Chen Y.Y., Du J.S., Chen Y.Y., Chung M.C., Tsai W.C., et al, 2014. Flower development of Phalaenopsis orchid involves functionally divergent sepallatalike genes. New Phytol, 202: 1024-1042. DOI:10.1111/nph.12723 |
Ramirez S.R., Gravendeel B., Singer R.B., Marshall C.R., Pierce N.E., 2007. Dating the origin of the Orchidaceae from a fossil orchid with its pollinator.. Nature, 448: 1042-1045. DOI:10.1038/nature06039 |
Schultz E.A., Haughn G.W., 1991. LEAFY, a homeotic gene that regulates inflorescence development in Arabidopsis.. Plant Cell, 3: 771-781. DOI:10.2307/3869271 |
Somssich M., Je B.I., Simon R., Jackson D., 2016. CLAVATA-WUSCHEL signaling in the shoot meristem.. Development, 143: 3238-3248. DOI:10.1242/dev.133645 |
Spinelli S.V., Martin A.P., Viola I.L., Gonzalez D.H., Palatnik J.F., 2011. A mechanistic link between STM and CUC1 during Arabidopsis development. Plant Physiol, 156: 1894-1904. DOI:10.1104/pp.111.177709 |
Su S., Shao X., Zhu C., Xu J., Lu H., Tang Y., Jiao K., Guo W., Xiao W., Liu Z., et al, 2018a. Transcriptome-wide analysis reveals the origin of peloria in Chinese Cymbidium (Cymbidium sinense). Plant Cell Physiol, 59: 2064-2074. DOI:10.1093/pcp/pcy130 |
Su S., Shao X., Zhu C., Xu J., Tang Y., Luo D., Huang X., 2018b. An AGAMOUS-like factor is associated with the origin of two domesticated varieties in Cymbidium sinense (Orchidaceae). Hortic.Res, 5: 48. DOI:10.1038/s41438-018-0052-z |
Sun Y., Wang G., Li Y., Jiang L., Yang Y., Guan S., 2016. De novo transcriptome sequencing and comparative analysis to discover genes relatChinese Cymbidiumed to floral development in Cymbidium faberi Rolfe.. SpringerPlus, 5: 1458. DOI:10.1186/s40064-016-3089-1 |
Tao Y., Chen M., Shu Y., Zhu Y., Wang S., Huang L., Yu X., Wang Z., Qian P., Gu W., et al, 2018. Identification and functional characterization of a novel BEL1-LIKE homeobox transcription factor GmBLH4 in soybean.. Plant Cell Tiss Org (PCTOC), 134: 331-344. DOI:10.1007/s11240-018-1419-4 |
Tsai W.C., Kuoh C.S., Chuang M.H., Chen W.H., Chen H.H., 2004. Four DEF-like MADS box genes displayed distinct floral morphogenetic roles in Phalaenopsis orchid. Plant Cell Physiol, 45: 831-844. DOI:10.1093/pcp/pch095 |
Uemura A., Yamaguchi N., Xu Y., Wee W., Ichihashi Y., Suzuki T., Shibata A., Shirasu K., Ito T., 2018. Regulation of floral meristem activity through the interaction of AGAMOUS, SUPERMAN, and CLAVATA3 in Arabidopsis. Plant Reprod, 31: 89-105. DOI:10.1007/s00497-017-0315-0 |
Wang J., Tian C., Zhang C., Shi B., Cao X., Zhang T.Q., et al, 2017. Cytokinin signaling activates wuschel expression during axillary meristem initiation.. Plant Cell, 29: 1373-1387. DOI:10.1105/tpc.16.00579 |
Xiang L., Chen Y., Chen L., Fu X., Zhao K., Zhang J., Sun C., 2018. B and E MADSbox genes determine the perianth formation in Cymbidium goeringii. Rchb.f.Physiol Plant, 162: 353-369. DOI:10.1111/ppl.12647 |
Yan L., Wang X., Liu H., Tian Y., Lian J., Yang R., Hao S., Yang S., Li Q., Qi S., et al, 2015. The genome of Dendrobium officinale illuminates the biology of the important traditional Chinese orchid herb.. Mol.Plant, 8: 922-934. DOI:10.1016/j.molp.2014.12.011 |
Yang F., Zhu G., 2015. Digital gene expression analysis based on de novo transcriptome assembly reveals new genes associated with floral organ differentiation of the orchid plant Cymbidium ensifolium.. PloS One, 10: e0142434. DOI:10.1371/journal.pone.0142434 |
Yang F., Zhu G., Wang Z., Liu H., Xu Q., Huang D., Zhao C., 2017. Integrated mRNA and microRNA transcriptome variations in the multi-tepal mutant provide insights into the floral patterning of the orchid Cymbidium goeringii. BMC Genom, 18: 367. DOI:10.1186/s12864-017-3756-9 |
Yang F., Zhu G., Wei Y., Gao J., Liang G., Peng L., Lu C., Jin J., 2019. Low-temperature-induced changes in the transcriptome reveal a major role of CgSVP genes in regulating flowering of Cymbidium goeringii. BMC Genom, 20: 53. DOI:10.1186/s12864-019-5425-7 |
Yu H., Ito T., Wellmer F., Meyerowitz E.M., 2004. Repression of AGAMOUS-LIKE 24 is a crucial step in promoting flower development. Nat.Genet, 36: 157-161. DOI:10.1038/ng1286 |
Zhang Z., Yan Y., Tian Y., Li J., He J.-S., Tang Z., 2015. Distribution and conservation of orchid species richness in China. Biol.Conserv, 181: 64-72. DOI:10.1016/j.biocon.2014.10.026 |
Zhang G.Q., Liu K.W., Li Z., Lohaus R., Hsiao Y.Y., Niu S.C., Wang J.Y., Lin Y.C., Xu Q., Chen L.J., et al, 2017. The Apostasia genome and the evolution of orchids.. Nature, 549: 379-383. DOI:10.1038/nature23897 |



