b. CAS Key Laboratory for Plant Diversity and Biogeography of East Asia, Kunming Institute of Botany, Chinese Academy of Sciences, Kunming 650201, China;
c. State Key Laboratory of Systematic and Evolutionary Botany, Institute of Botany, Chinese Academy of Sciences, Beijing 100093, China;
d. College of Biology and Environmental Sciences, Jishou University, Jishou 416000, China
Crepis L. is a large taxonomically controversial genus in tribe Cichorieae of Asteraceae containing over 200 species that are mainly distributed throughout the Northern Hemisphere and Africa (Enke and Gemeinholzer, 2008; Shi et al., 2011). The genus presumably originated in the Altai/Tien Shan region in Central Asia (Babcock, 1947). The present center of diversity of Crepis is the circum-Mediterranean area (Enke and Gemeinholzer, 2008). Babcock, 1947 divided Crepis into 27 sections worldwide and included Lagoseris as a section of the genus Crepis (e.g., Lack, 2007; Enke, 2010). However, recent molecular studies based on nuclear ITS and plastid matK sequences indicated that sect. Lagoseris may be a distinct genus (Enke and Gemeinholzer, 2008; Enke, 2010; Kilian et al., 2009). ITS and matK phylogenetic trees of the Lagoseris clade include Crepis species from sections Lagoseris, Pterotheca, Microcephalum, Intybellia, as well as the genera Rhagadiolus and Lapsana (Enke and Gemeinholzer, 2008; Enke, 2010).
During fieldwork (2012-2013) in a sand desert area of the Junggar Basin of northern Xinjiang in northwestern China, we discovered an ephemeral species of Asteraceae that could not be identified. Its annual habit and the receptacle with only a few setiform paleae indicated that the plant clearly was a member of Crepis sect. Pterotheca (Babcock, 1947a, 1947b) or Lagoseris subg. Pterotheca (Czerepanov, 1964). Detailed examination of these plants and a survey of the literature, including Flora of the U.S.S.R (Czerepanov, 1964), Flora of China (Shi et al., 2011), Flora Xinjiangensis (An, 1999) and Babcock's monographs of Crepis (Babcock, 1947a, 1947b), showed that this unknown plant was similar to Crepis sancta subsp. bifida (Vis.) Thell. ex Babc. but distinguished from it by glandular hairs on the stem, bract shape, style color and shape of the dimorphic achenes. Morphological comparisons and nuclear ITS and plastid matK sequences indicated that this plant represented an undescribed species of Crepis, which we named Crepis desertorum.
2. Material and methods 2.1. Morphological descriptionThe morphological description is based on live material. Details of bracts, flowers and achenes were examined under a Nikon SMZ1000 stereoscopic microscope (Nikon Corp., Tokyo, Japan) and measured with a digital caliper. Distribution, habitats and phenology of the new species are based on field observations. Live material was collected from Mosuowan, Shihezi city of Xinjiang, China (44°25.42'N, 86°01.21'E).
2.2. SEM observations on pollen and achene morphologyMature pollen grains and achenes were mounted directly on aluminum stubs with double-coated conductive glue, coated with gold-palladium and examined under a Zeiss SUPRA 55VP (Carl Zeiss, Oberkochen, Germany) scanning electron microscope (SEM). Terminology for the description of pollen follows Skvarla and Turner (1966) and Ma et al. (2009). Vouchers for the pollen and achene material studied are provided in Table 1. All voucher specimens have been deposited in the Herbarium of Xinjiang Agricultural University (XJA) (see Table 1).
| Location | Coordinate | Altitude | Date | Voucher | |
| Pollen material |
China, Xinjiang, Shihezi City Mosuowan District, 147 Regiment 22 Company | 44°25.420N, 86°01.210E |
300 m | 6 May, 2013 | J. Qiu 2013-0171 (XJA) |
| China, Xinjiang, Jimsar County, Wucaiwan District, China national highway 216 514 km | 44°23.710N, 88° 47.210E |
530 m | 27 Apr, 2013 | J. Qiu 2013-0129 (XJA) |
|
| Achene material |
China, Xinjiang, Shihezi City Mosuowan District, 147 Regiment 22 Company | 44°25.420N, 86°01.210E |
300 m | 15 May, 2013 | J. Qiu 2013-0195 (XJA) |
| China, Xinjiang, Jimsar County, Wucaiwan District, China national highway 216 514 km | 44°23.710N, 88°47.210E |
530 m | 12 May, 2013 | D.Y. Tan 274 (XJA) |
Mature achenes were collected from a natural population (Mosuowan, Shihezi city of Xinjiang, China) in June 2013. They were incubated on wet filter paper in Petri dishes at 20/10 ℃ (12/12 h) and a 12 h dailylight period for 4-5 days, during which time germination occurred. Root-tips were removed from roots 1-1.5 cm in length and pretreated with 0.002 M 8-hydroxyquinoline at room temperature (c. 20 ℃) for 3 h and then fixed in ethanol: glacial acetic acid (3:1v/v) for 24 h. Subsequently, fixed root-tips were hydrolyzed in 1 M HCl for 20 min at 60 ℃, stained with carbol fuchsin and then squashed in a drop of 45% acetic acid and mounted for cytological observations. Photomicrographs were taken under a Zeiss Axioskop 40 microscope (Carl Zeiss, Oberkochen, Germany) using a 100 × oil lens. Karyotypic analysis of chromosomes at mitotic-metaphase was determined from five well-spread metaphases. The symbols used to describe the karyotypes followed Levan et al. (1964) and Li and Chen (1985). The asymmetry classification of the karyotype follows Stebbins (1971) and Li and Chen (1985).
2.4. Molecular analysisWe sampled 17 populations representing 14 species of Crepis s.l., including the presumed new species, for molecular analyses. Askellia flexuosa was chosen for the outgroup based on Enke and Gemeinholzer (2008), Zhang et al. (2011), and our data. Sequences for other related taxa were obtained from GenBank (see Appendix S1). A list of samples, voucher information and GenBank accession numbers are provided in Appendix S1.
DNA was extracted from leaves dried over silica-gel using the CTAB method (Doyle and Doyle, 1987). DNA sequences of the chloroplast matK region and of nuclear ribosome internal transcribed spacers (ITS) were amplified based on a PCR procedure of initial denaturation at 94 ℃ for 2 min, followed by 40 cycles of 40 s at 94 ℃, 1 min 30 s at 50 ℃ and 2 min at 72 ℃, with a final extension at 72 ℃ for 15 min. Amplification products were purified on 1.5% low-melting agarose gels, and the desired DNA was recovered with a UNIQ-10 kit (Shanghai SBS, Biotech Ltd., China), following directions provided by the manufacturer. Sequencing reactions were conducted with the forward or reverse primers of the amplification reactions, using the DYEnamic ET Terminator Kit (Amersham Pharmacia Biotech, Shanghai, China), with an ABIPRISM3730 automatic DNA sequencer from Shanghai Sangon Biological Engineering Technology & Services Co., Ltd. (Shanghai, China). The PCR primers for the nuclear internal transcribed spacers (ITS) were ITS4 and ITS5 (White et al., 1990), and the chloroplast matK sequences were amplified in two overlapping parts using the primers trnK-710f (Johnson and Soltis, 1995) and matK-iR (Fehrer et al., 2007) for matK 1, matK-ifN and matK-rN (Enke and Gemeinholzer, 2008) for matK 2.
The program Sequencher 4.1 (Gene Codes Corp., Ann Arbor, Michigan, USA) was used to evaluate chromatograms for base confirmation and to edit contiguous sequences. After editing, all sequences were aligned in MAFFT v7.311 (Katoh et al., 2002; Katoh and Standley, 2013) and manually adjusted with MEGA 7.0.14 (Kumar et al., 2016). Phylogenetic analyses were conducted based on Bayesian inference (BI) in MrBayes v3.2.6 (Ronquist et al., 2012), Maximum Likelihood in RAxML 8.2.10 (Stamatakis, 2014) on CIPRES (Miller et al., 2010) and Maximum Parsimony (MP) in PAUP version 4.0b10 (Swofford, 2002).
For Bayesian inference (BI) analysis, jModeltest v2.1.7 (Darriba et al., 2012) was used to select the best-fit model of nucleotide substitution based on the Akaike information criterion (AIC). The SYM + I + G model was selected for the nrITS dataset and the GTR + I model for the cpDNA dataset. Four simultaneous Monte Carlo Markov chains (MCMCs) were run for five million generations for both nrITS, matK and combined sequences, and one tree was sampled every 1000 generations. The first 1250 trees (25% of total trees) were discarded as burn-in. The remaining trees were summarized in a 50% majority-rule consensus tree, and the Bayesian posterior probabilities (BP) were calculated.
For the maximum likelihood bootstrap (ML) analyses, we constructed the phylogeny using GTRCAT as the nucleotide substitution model and performed 1000 replicates for the bootstrap analyses.
For the maximum parsimony bootstrap (MP) analyses, 100 replicates of random stepwise addition with tree bisection-reconnection (TBR) branch swapping were performed using heuristic searches, with all most-parsimonious trees saved at each replicate (MulTree on). Support for each branch was assessed using bootstrap analyses with 100 bootstrap replicates, each with 10 stepwise additions.
3. Results 3.1. Taxonomic treatmentCrepis desertorum J. Qiu & D.Y. Tan, sp. nov (Figs. 1 and 2).

|
| Fig. 1 Illustrations of Crepis desertorum J. Qiu & D.Y. Tan sp.nov. Drawn by C.D. Cui. (A) individual; (B) capitulum; (C) outer bract; (C′) inner bract; (D) floret; (E) ligule teeth; (F) receptacle; (G) infructescence; (H) peripheral achene; (H′) central achene. |
|
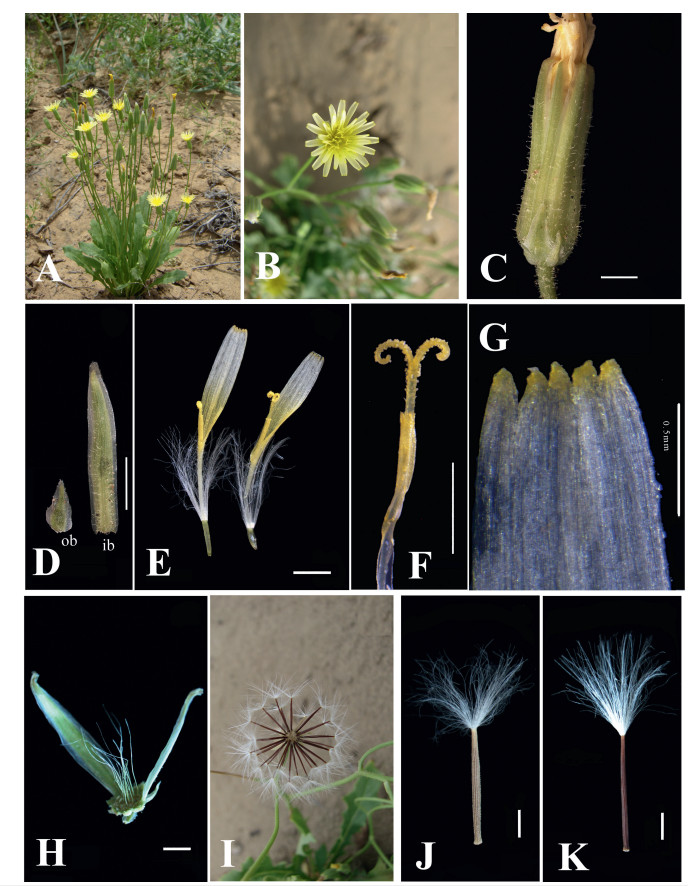
| Fig. 2 Photographs of Crepis desertorum J. Qiu & D.Y. Tan. (A) individual; (B) capitulum; (C) involucre; (D) bracts (ob = outer bracts, ib = inner bracts); (E) ligulate florets; (F) style branches; (G) details of ligule teeth; (H) receptacle; (I) infructescence; (J) peripheral achene; (K) central achene. Scale bar: 1 mm. |
Type: CHINA. Xinjiang, Mosuowan District, Shihezi city, sand dune, 44°25.42'N, 86°01.21'E, alt. 300 m, 30 Apr. 2013, J. Qiu & D. Y. Tan 2013-0146 (holotype: XJA, isotypes: KUN and PE).
3.2. DiagnosisC. desertorum is similar to C. sancta subsp. bifida, but it is easily distinguished from the latter species by habitat, stem with glandular hairs, style color, receptacle and dimorphic achenes.
3.3. DescriptionAnnual. Roots slender, hard. Stem erect or ascending, 7-25 cm high, cylindrical with glandular hairs, finely ribbed. Basal leaves numerous and forming a rosette, leaf blade linear-lanceolate, lanceolate or oblanceolate, 4-10 cm long (including petiole) and 0.7-1.4 cm wide, glabrous, base attenuate to petioles, apex obtuse to acute, margin irregularly sinuate-dentate or pinnatipartite, spiny hairy marginally; cauline leaves arising from the branches, linear, ca. 0.3-0.4 mm long. Capitula many in corymbs. Involucre narrowly cylindrical to narrowly campanulate, 7-11 mm long and 2-4 mm wide. Involucral bracts with glandular hairs; outer bracts 4-5, triangular, longest 1/3-1/4 as long as inner ones, margin white scarious, apex acute; inner bracts 8-12, lanceolate, 10-12 mm long, ca. 2 mm wide, margin white scarious, apex acute. Receptacle covered with erect setaceous hairs, 3-7 mm long. Florets ligulate, 12-30, corolla 6-9 mm long, tube white, ca. 3 mm long, ligule yellow in lower part and yellowish-white in upper part, apex 5- dentate with teeth ca. 0.01 mm long; anther ca. 2 (-2.5) mm long; style branches yellowish, 0.5 mm long. Infructescences numerous. Achenes dimorphic; peripheral achenes 2-15, whitish-yellowish, terete, linear, 4.2-6.1 mm long and 0.2-0.5 mm wide, with 10 ribs covered with papilliform protuberances; central achenes 7-30, reddish-brown, terete, linear, 4.2-5.6 mm long and 0.2-0.4 mm wide, with 10 smooth ribs. Pappus many fine white bristles, 3-6 mm long. Fl. and fr. Apr.-Jun.
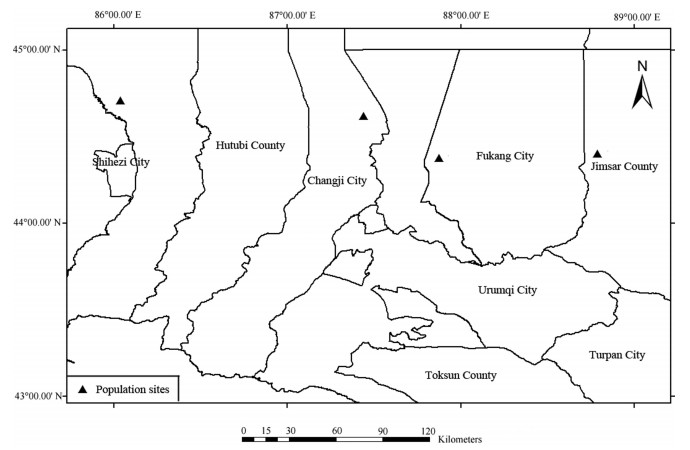
Distribution and habitat - C. desertorum is widely distributed in the Junggar Basin. It grows in stationary sand dunes at altitudes of 300-530 m (Fig. 5). Associated species include Haloxylon ammodendron (C.A.Mey.) Bunge, Calligonum leucocladum (Schrenk) Bge., Eremurus anisopterus (Kar. et Kir.) Regel, E. inderiensis (M. Bieb.) Regel, Soranthus meyeri Ledeb., Astragalus flexus Fisch. and some annual desert ephemerals.
Etymology - The specific epithet is derived from the desert habitat, from which the type of this species was collected.
Chinese name - Huang Mo Huan Yang Shen (荒漠还阳参).
Morphological evidence - By comparing the morphological characteristics of C. desertorum with those of its related species (Table 3), we found that C. desertorum is similar to the species C. sancta (with three subspecies) and more specifically to C. sancta subsp. bifida. However, it differs from C.sancta in having glandular hairs on the stem (Fig. 1A) (vs. yellow setiform hairs, subglabrous or glandular on the stem of C. sancta), a flat receptacle with some erect setaceous hairs in the center of the receptacle (Figs. 1F and 2H) (vs. flat receptacle with scales in C. sancta), style usually yellow (vs. green in C. sancta subsp. bifida) and dimorphic achenes (vs.trimorphic achenes in C.sancta and C.sancta subsp.bifida). The relatedspecies C. frigida is a perennial with monomorphic achenes (Table 3), whereas C. desertorum is an annual herb usually with dimorphic achene (Figs. 1H, H' and 2J, K).
| Chromosome pair | Relative length | Centromeric index (CI) | Arm ratio (AR) | Chromosome types | ||
| L | S | TLC | ||||
| 1* | 3.86 | 1.21 | 5.06 | 22.86 | 3.20 | st |
| 2 | 3.44 | 1.61 | 5.04 | 31.96 | 2.14 | sm |
| 3 | 3.39 | 1.21 | 4.60 | 25.92 | 2.81 | sm |
| 4 | 2.36 | 1.57 | 3.94 | 39.94 | 1.50 | m |
| 5 | 2.26 | 1.37 | 3.63 | 38.01 | 1.65 | m |
| Note: * Chromosomes with satellites. st (subtelocentric), sm (submetacentric) and m (metacentric). | ||||||
| Characters | Crepis desertorum | Crepis frigidaa'b | Crepis sanctaa | Crepis sancta subsp. bifidac |
| Life form | annual | perennial | annual | annual |
| Stem indumentum | glandular hairs | mostly scapelike, tomentose and pubescent | yellow setiform hairs, subglabrous or glandular | — |
| Outer bracts | triangular, longest 1/3—1/4 as long as inner ones, c. 0.3 —0.4 mm long, margin white scarious, apex acute | narrowly ovate, acute | linear to ovate, 2.0—3.0 mm long, c. 1.5 mm wide, subglabrous or thinly glandular, margin white scarious | deltoid to subovate, acute, 1/8-1/3 as long as the inner bracts |
| Receptacle | flat, covered with erect setaceous hairs | flat, with scales | flat, with scales | — |
| Style color | yellow | yellow | greenish or yellow | green |
| Achene type | dimorphic | monomorphic | dimorphic or sometimes trimorphic | dimorphic or sometimes trimorphic |
| Peripheral achenes, shape | whitish-yellowish, terete, linear, 4.2—6.1 mm long and 0.2—0.5 mm wide, with 10 ribs covered with dense papilliform protuberances | light brown, cylindrical, c. 5.0 mm long, straight or curved, with 15 smooth ribs, slightly attenuate; epidermal cells obscure in outline with acute end wall, outer cell wall ornamentation smooth | white, cylindrical, 3.0—5.0 mm long, 0.6—1.5 mm wide, sulcate or smooth dorsally and mostly winged, stramineous (sometimes absent) | white, corticeous, strongly obcompressed, linear-lanceolate, dorsally smooth, the lateral alae narrow, thin and indurate (sometimes absent) |
| Intermediate achenes, shape | — | — | terete, slender, coarsely or finely spiculate | terete, slender, coarsely or finely spiculate |
| Central achenes, shape | reddish-brown, linear, 4.2 —5.6 mm long and 0.2 —0.4 mm wide, with 10 smooth ribs; the epidermal cells obscure in outline with acute end wall | - | brown or greenish, narrowly fusiform, 3.0—4.0 mm long, c. 0.4 mm wide, sometimes scabridulous | terete, slender, smooth |
| Note: -, Not described. a http://cichorieae.e-taxonomy.net/portal/node/8. b c |
||||
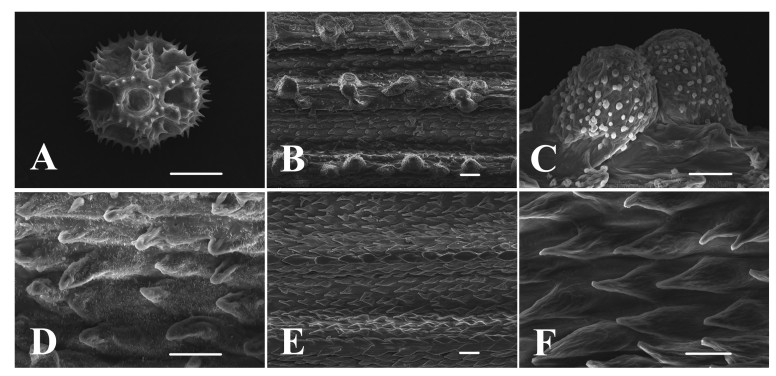
Pollen evidence - C. desertorum pollen is echinoloplate and belongs to the Cichorium intybus type. It is isopolar, radially symmetrical, tricolporate and spheroidal. Grain size ranges from 28.21 to 35.48 mm (Fig. 3A). The endexine (a distinct exine layer between ektexine and intine) is thick, and the columellae are very thin and short.
|
| Fig. 3 SEM images of pollen and achenes of Crepis desertorum J. Qiu & D.Y. Tan. (A) pollen grain (polar view); (B-D) Peripheral achene (B) rough longitudinal ribs; (C) papilliform cells covered with white granules; (D) scaly cells covered with short tomentum; (E-F) Central achene (E) thin longitudinal ribs; (F) glabrous scaly cell. Scale bar: 10 μm in A, C, D and F, 20 μm in B and E. |
Achene morphology - Peripheral and central achenes of C. desertorum differ in pericarp micro-morphology. Rough longitudinal ribs (Fig. 3B) on the surface of peripheral achenes are composed of papilliform cells covered with white granules (Fig. 3C). Epidermal cells between the longitudinal ribs are scaly and covered with short tomentum (Fig. 3D). However, thin longitudinal ribs (Fig. 3E) on the surface of central achenes are composed of glabrous scaly cells (Fig. 3F), and the epidermal cells between the longitudinal ribs also are scaly and glabrous.
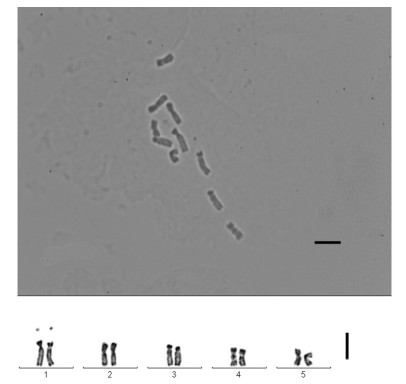
Karyotype evidence - The chromosome number of C. desertorum is 2n = 2x = 10 (Fig. 4). The karyotype includes 5 pairs of chromosomes (Table 2): pair 1 subtelocentric (st), pairs 2-3 submetacentric (sm), and pairs 4-5 metacentric (m). The first pair has a satellite. The karyotype formula is 2n = 10 = 4m + 4sm + 2nd (2SAT). Karyotype asymmetry is of the 3A type.
|
| Fig. 4 Metaphase chromosomes and karyotype of Crepis desertorum J. Qiu & D.Y. Tan. Scale bar: 5 μm. |
|
| Fig. 5 Distribution of Crepis desertorum J. Qiu & D.Y. Tan in the Junggar Basin of Xinjiang, China. Mapped by H. F. Zhang. |
Phylogenetic evidence - The Bayesian tree with MP, ML and BP support values for each clade is shown in Fig. 6. The sampled species of Crepis s.l. form a monophyletic group with strong support of BP = 1.00, when the genera Lapsana and Rhagadiolus, which are deeply nested within the clade of Crepis s.l., are included. The clade of Lapsana and Rhagadiolus is sister to the Lagoseris clade, which includes the new species C. desertorum and some species of the C. sect. Lagoseris (C. frigida, C. sancta, C. purpurea, C. sahendi, C. multicaulis, C. elongata). Within the clade, C. desertorum is sister to C. frigida but with moderate support of BP = 0.78.
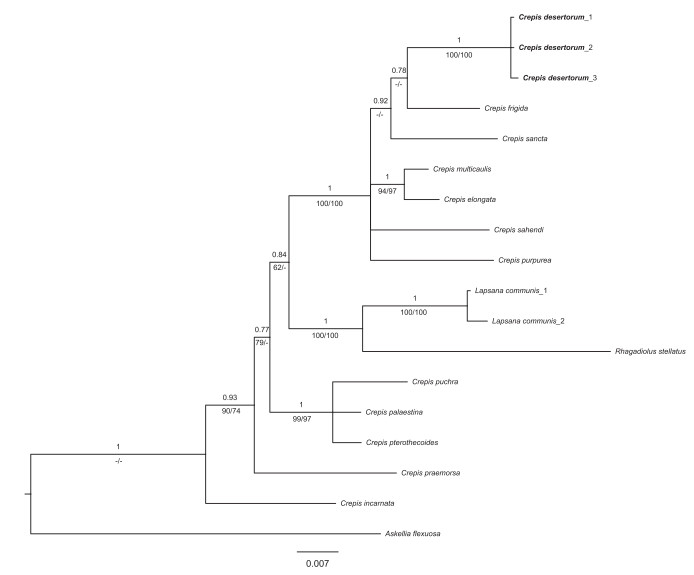
|
| Fig. 6 Bayesian consensus tree of Crepis desertorum (bold) and related species of Crepis. Numbers above branches indicate Bayesian posterior probability (BP), numbers below branches are maximum likelihood bootstrap (ML) and maximum parsimony bootstrap (MP). |
Paratypes - CHINA. Xinjiang: Mosuowan District, Shihezi City, alt. 300 m, 12 June, 2012, D. Y. Tan 2012-0102 (XJA); same locality, 6 May, 2013, J. Qiu 2013-0171 (XJA); Wujiaqu City, alt. 490 m, 12 May, 2013, J. Qiu 2013-0190 (XJA); Wutonggou, Fukang City, alt. 446 m, 27 Apr., 2013, J. Qiu 2013-0142 (XJA); same locality, 14 May, 2013, J. Qiu 2013-0182 (XJA); Wucaiwan District, Jimsar County, alt. 530 m, 27 Apr., 2013, J. Qiu 2013-0093, 2013-0129 (XJA).
4. DiscussionThe genus Crepis comprises about 18 species (five endemic) in China, nine of which are distributed in Xinjiang (Shi et al., 2011). The Lagoseris species previously reported as occurring in Xinjiang is L. sancta (L.) K. Maly (Hou, 1982). L. sancta also is recorded in Flora Xinjiangensis (An, 1999). However, neither Hou (1982) nor An (1999) described the distribution or habitat of L. sancta in Xinjiang. Moreover, we did not find any voucher specimen in the herbaria of the Xinjiang Institution of Ecology and Geography, Chinese Academy of Sciences (XJBI), Shihezi University (SHI) or Xinjiang Agricultural University (XJA). In addition, no relevant records of specimens were found in the Chinese virtual herbarium (http://www.cvh.ac.cn/) or in the Specimen resources sharing platform for education of China (http://mnh.scu.edu.cn/main.aspx).
The pollen of C. desertorum and C. sancta belongs to the Cichorium intybus type, but the pollen of C. desertorum belongs to the C. intybus subgroup, while that of C. sancta belongs to the Taraxacum officinalis subgroup (Enke, 2010). However, C. desertorum has a chromosome number of x = 5, which differs from that of C. frigida (x = 4) (Enke and Gemeinholzer, 2008).
Molecular studies based on nuclear ITS and plastid matK sequences indicated that C. desertorum is more closely related to C. frigida than to C. sancta subsp. bifida (Fig. 6) but with low support (PP = 0.78). Although the micromorphology of the central achenes of C. desertorum is similar to that of C. frigida (Kalmuk et al., 2018), the two species differ greatly in life form, stem indumentum and outer bracts (Table 3). Babcock, 1947a, 1947b even treated L. sancta as a synonym of C. sancta subsp. bifida. C. desertorum is known only from the Junggar Basin in northern Xinjiang, China; C. sancta is distributed in Europe, the Caucasus region, Balkans-Asia Minor, Palestine, Afghanistan and the Indo-Himalayas, whereas C. sancta subsp. bifida is distributed in Afghanistan, Turkestan and N.W. Himalayas (Babcock, 1947; Czerepanov, 1964). Thus, the geographical distribution of C. desertorum is disjunct from that of C. sancta and C. sancta subsp. bifida. C. desertorum has some unique morphological characters, and it differs from C. sancta subsp. bifida in having glandular hairs on the stem, yellowish style branches, peripheral whitish-yellowish linear achenes with 10 ribs covered with papilliform protuberances and white pappus (Table 3).
Africa is a major distribution area of the genus Crepis with more than 40 species, and the achenes of some African species are dimorphic, such as C. aspera, C. amplexifolia, C. pulchra, and others (http://cichorieae.e-taxonomy.net/portal/node/8). There are significant differences in the achene shape and number of the ribs, surface structure and the pappus between C. desertorum and these African species with dimorphic achenes (Enke, 2010; Kalmuk et al., 2018; http://cichorieae.e-taxonomy.net/portal/node/8).
Declaration of Competing InterestThe author declares no conflict of interest.
AcknowledgementsWe thank Drs. Carol C. Baskin and Jerry M. Baskin (University of Kentucky, Lexington, Kentucky, USA) for correcting the English, and we are grateful to Dr. H. L. Chen for molecular lab work and analysis. This study was supported by the National Natural Science Foundation of China (No. U1603231, No. 31650067 and No. 31570213) and Program for Tianshan Innovative Research Team of Xinjiang Uygur Autonomous Region, China (2018D14010).
Appendix S1Taxon name, voucher information (for the first published sequences in part I and the downloaded sequences from GenBank that was placed in part II) and GenBank accession numbers for ITS and matK sequences used in this study.
| Taxa | Voucher | ITS | matK |
| Crepis desertorum J. Qiu & D.Y. Tan_1 |
LW-079 (XJA), Wucaiwan, | MN750316 | MN764299 |
| Crepis desertorum J. Qiu & D.Y. Tan_2 |
Xinjiang, China | MN750317 | MN764300 |
| Crepis desertorum J. Qiu & D.Y. Tan_3 |
MN750318 | MN764301 | |
| Crepis elongata Babcock | SunH2522 (KUN), Demula, Tibet, China | MN750319- | MN764302 |
| Taxa | ITS | matK |
| Askellia flexuosa (Ledebour) W. A. Weber | EU363596 | EU363544 |
| Crepis frigida (Boiss. & Balansa) Babcock | EU363612 | EU363555 |
| Crepis rubra L. (= C. incarnata Vis.) | EU363608 | EU363551 |
| Crepis multicaulis Ledeb. | EU363642 | EU363573 |
| Crepis palaestina (Boiss.) Bornm. | EU363639 | - |
| Crepis praemorsa (L.) Walther | EU363654 | EU363578 |
| Crepis pteothecoides Boiss. | - | EU363570 |
| Crepis pulchra L. | AJ633369 | AJ633144 |
| Crepis purpurea (Willd.) M. Bieb. | EU363653 | - |
| Crepis sahendi Boiss. & Buhse | EU363651 | - |
| Crepis sancta (L.) Bornm. | AJ633372 | AJ633150 |
| Lapsana communis L._1 | AJ633285 | AJ633137 |
| Lapsana communis L._2 | AJ633286 | AJ633138 |
| Rhagadiolus stellatus (L.) Gaertn. | DQ451823 | AJ633224 |
An, Z.X., 1999. Cichorieae. In: An, Z.X. (Ed.), Flora Xinjiangensis, vol. 5. Xinjiang Science & Technology & Hygiene Publishing House, Urumqi, pp. 367-472.
|
Babcock, E.B., 1947a. The Genus Crepis I. The Taxonomy, Phylogeny, Distribution and Evolution of Crepis, 21. University of California Publications in Botany, pp. 1-198.
|
Babcock, E.B., 1947b. The Genus Crepis II. Systematic Treatment, 22. University of California Publications in Botany, pp. 199-1030.
|
Czerepanov, S.K., 1964. Lagoseris. In: Bobrov, E.G., Tzvelev, N.N. (Eds.), Flora of the USSR, 29. Academy of Sciences of the USSR, Moscow & Leningrad, pp. 699-715.
|
Darriba D., Taboada G.L., Doallo R., Posada D., 2012. jModelTest 2: more models, new heuristics and parallel computing. Nat.Methods, 9(8): 772. DOI:10.1038/nmeth.2109 |
Doyle J.J., Doyle J.L., 1987. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem.Bull, 19(1): 11-15. |
Enke N., 2010. Contributions towards a revised infrageneric classification of Crepis(Cichorieae, Compositae). Willdenowia, 39(2): 229-245. DOI:10.3372/wi.39.3920 |
Enke N., Gemeinholzer B., 2008. Babcock revisited: new insights into generic delimitation and character evolution in Crepis L.(Compositae: Cichorieae) from ITS and matK sequence data.. Taxon, 57(3): 756-768. DOI:10.1002/tax.573008 |
Fehrer J., Gemeinholzer B., Chrtek J., Br#228;utigam S., 2007. Incongruent plastid and nuclear DNA phylogenies reveal ancient intergeneric hybridization in Pilosella hawkweeds (Hieracium Cichorieae, Asteraceae). Mol.Phylogenetics Evol, 42(2): 347-361. DOI:10.1016/j.ympev.2006.07.004 |
Hou, K.Z., 1982. A Dictionary of the Families and Genera of Chinese Seed Plants, second ed. Science Press, Beijing, p. 263.
|
Johnson L.A., Soltis D.E., 1995. Phylogenetic inference in Saxifragaceae sensu stricto and Gilia (Polemoniaceae) using matK sequences. Ann.Mo.Bot.Gard, 82(2): 149-175. DOI:10.2307/2399875 |
Kalmuk N.A., Inceer H., Imamoglu K.V., 2018. Achene micromorphology of 26 Crepis L.(Asteraceae) taxa from Turkey with notes on its systematic and ecological significance.. Bot.Lett, 165: 292-306. DOI:10.1080/23818107.2018.1452167 |
Katoh K., Misawa K., Kuma K., Miyata T., 2002. MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res, 30(4): 3059-3066. DOI:10.1093/nar/gkf436 |
Katoh K., Standley D.M., 2013. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol.Biol.Evol, 30(4): 772-780. DOI:10.1093/molbev/mst010 |
Kilian, N., Gemeinholzer, B., Lack, H.W., 2009. Cichorieae. In: Funk, V.A., Susana, A., Stuessy, T.F., Bayer, R.J. (Eds.), Systematics, Evolution and Biogeography of the Compositae. IAPT, Vienna, pp. 343-383.
|
Kumar S., Stecher G., Tamura K., 2016. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets.. Mol.Biol.Evol, 33(7): 1870-1874. DOI:10.1093/molbev/msw054 |
Lack, H.W., 2007. Tribe Cichorieae Lam. et DC. In: Kadereit, J.M., Jeffrey, C. (Eds.), The Families and Genera of Vascular Plants, vol. 8. Springer, Berlin & Heidelberg, pp. 180-199.
|
Levan A., Fredga K., Sandberg A.A., 1964. Nomenclature for the centromeric position on chromosomes.. Hereditas, 52(2): 201-220. |
Li M.X., Chen R.Y., 1985. A suggestion on the standardization of karyotype analysis in plants. J.Wuhan Bot.Res, 3(4): 297-302. |
Ma Y.Z., Meng H.W., Sang Y.L., Sun A.Z., Wu J., Wang W., 2009. Pollen keys for identification of coniferopsida and compositae classes under light microscopy and their ecological significance. Acta Palaeontol.Sin, 48(2): 240-253. |
Miller, M.A., Pfeiffer, W., Schwartz, T., 2010. Creating the CIPRES Science Gateway for inference of large phylogenetic trees. In: Proceedings of the Gateway Computing Environments Workshop (GCE). New Orleans, LA, pp. 1-8.
|
Ronquist F., Teslenko M., van der Mark P., Ayres D.L., Darling A., H#246;hna S., Larget B., Liu L., Suchard M.A., Huelsenbeck J.P., 2012. MrBayes 3.2: efficient bayesian phylogenetic inference and model choice across a large model space.. Syst.Biol, 61(3): 539-542. DOI:10.1093/sysbio/sys029 |
Shi, Z., Ge, X.J., Kilian, N., Kirschner, J., Štěpánek, J., Sukhorukov, A.P., Mavrodiev, E.V., Gottschlich, G., 2011. Cichorieae. In: Wu, Z.Y., Raven, P.H., Hong, D.Y. (Eds.), Flora of China, vol. 20. Science Press, Beijing & Missouri Botanical Garden Press, St. Louis, pp. 195-353.
|
Skvarla J.J., Turner B.L., 1966. Systematic implications from electron microscopic studies of Compositae pollen e a review. Ann.Mo.Bot.Gard, 53(2): 220-256. DOI:10.2307/2394944 |
Stebbins, G.L., 1971. Chromosomal Evolution in Higher Plants. Edward Arnold Ltd., London, pp. 87-90.
|
Stamatakis A., 2014. RAxML version 8: a tool for phylogenetic analysis and postanalysis of large phylogenies. Bioinformatics, 30(9): 1312-1313. DOI:10.1093/bioinformatics/btu033 |
Swofford, D.L., 2002. PAUP*: Phylogenetic Analysis Using Parsimony (*and Other Methods). Sinauer, Sunderland, Massachusetts version 4.0b10.
|
White, T.L., Bruns, T., Lee, S., Taylor, J., 1990. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis, M.A., Gelfand, D., Sninsky, J.J., White, T.J. (Eds.), PCR Protocols: A Guide to Methods and Applications. Academic Press, San Diego, pp. 315-322.
|
Zhang J.W., Nie Z.L., Wen J., Sun H., 2011. Molecular phylogeny and biogeography of three closely related genera, Soroseris, Stebbinsia, and Syncalathium (Asteraceae, Cichorieae), endemic to the Tibetan Plateau, SW China. Taxon, 60(1): 15-26. DOI:10.1002/tax.601003 |



