b. University of Chinese Academy of Sciences, Beijing 100049, China;
c. Beijing Botanical Garden, Institute of Botany, Chinese Academy of Sciences, Beijing 100093, China
Biogeographical regionalization aims to classify global or regional biota into meaningful geographical units for analysis (Mackey et al., 2007). Thus, it can provide an indispensable background for research on biodiversity and conservation (Crisp et al., 2009). Takhtajan (1978) divided the global flora into six kingdoms, 34 regions and 147 provinces, which are widely used in historical and ecological biogeography (Linder et al., 2012; Meyer et al., 2012a, 2012b).
The increasing availability of species distribution data, combined with the tree of life reconstructions, have set new dimensions for studying biogeographical regionalization (Kreft and Jetz, 2010; Holt et al., 2013; Li et al., 2015, 2018; Li and Sun, 2017). Recently, important progress has been made in biogeographical regionalization using both qualitative approaches with endemic taxa (Wu et al., 2010) and quantitative approach with distribution and evolutionary data (Zhang et al., 2016a, 2016b; Ye et al., 2019). Compared with qualitative approaches, regionalization based on quantitative approaches are much more objective and reproducible. To arrive at a reasonable quantitative biogeographical regionalization scheme, it is critical to choose an appropriate dissimilarity index that measures pairwise distances between grid cells assemblages. Several metrics have been used in biogeographical regionalization, including Euclidean distance (Zhang et al., 2016a), Jaccard (Linder et al., 2005), the mean nearest taxon distance (MNTD) (Webb, 2000), Sørensen (Xie et al., 2004) and the turnover component of the Sørensen dissimilarity index (βsim; Baselga, 2012) (see Table 1). Some of these metrics are independent of differences in species richness among grid cells (Baselga, 2012), while some are strongly affected by differences in species richness (Lennon et al., 2001). For instance, Slik et al. (2018) suggested that the MNTD is easily affected by differences in taxonomic richness, and cannot give the correct measure of taxon turnover between two assemblages. Therefore, in studies of biogeographical regionalization, researchers must use those metrics (e.g., βsim and βjtu) that are least affected by variation in richness (Leprieur and Oikonomou, 2014), especially for those biotas with a large richness gradient (Mouillot et al., 2013), such as the Chinese flora (Lu et al., 2018).
| Reference | Taxon | Geographical extent | Dissimilarity index |
| Linder et al. (2012) | plants and vertebrates | sub-Saharan Africa | βsim |
| González-Orozco et al. (2013) | the genus Acacia | Australia | βsim |
| Holt et al. (2013) | amphibians, birds, and mammals | global | βsim |
| Mouillot et al. (2013) | fishes | Indo-Pacific | βsim and βjtu |
| Rueda et al. (2013) | birds, mammals and amphibians | global | Hellinger distance |
| González-Orozco et al. (2014a) | plants | Australia | βsim |
| González-Orozco et al. (2014b) | eucalypts | Australia and Malesia | βsim |
| Kubota et al. (2014) | woody plants | the Japanese archipelago | βsor |
| Hattab et al. (2015) | fishes | Mediterranean Sea | βjtu |
| Jønsson and Holt (2015) | passerine birds | global | βsor |
| Li et al. (2015) | plants | Yunnan, China | Bray—Curtis index, mean nearest |
| phylogenetic distance | |||
| Daru et al. (2016) | woody plants | southern Africa | βsim and βjtu |
| Julien et al. (2016) | orchids | New Guinea | βjac |
| Zhang et al. (2016a) | plants | Qinghai-Tibetan Plateau | Euclidean distance |
| Zhang et al. (2016b) | plants | China | βsim |
| Daru et al. (2017) | marine plants | global | βsim |
| He et al. (2017) | terrestrial vertebrates | China | βsim |
| König et al. (2017) | plants | global | βsim |
| Clark et al. (2017) | archaea | western Europe, the Mediterranean | βsim |
| and east Africa | |||
| Droissart et al. (2018) | plants | tropical Africa | Bray—Curtis index |
| Godinho and da Silva (2018) | anurans | Amazon | βsim |
| Slik et al. (2018) | plants | global | mean pairwise phylogenetic distance |
| Silva and Souza (2018) | plants | Caatinga | βsim |
| Bribiesca-Contreras et al. (2019) | brittle stars | global | βsim |
| Ye et al. (2019) | plants | China | βsim |
Because some previous studies used unreasonable metrics, in this study, we first explored the effect of metric choice on biogeographical regionalization, then compared the regionalizations of the Chinese flora resulting from different metrics by using species distribution records and a genus level phylogenetic tree of the Chinese flora. We then analyzed the boundaries, the characteristics, and origins of the chosen regions of the Chinese flora. We anticipate this work will help identify reasonable metrics that can be used to propose a phylogenetic floristic regionalization of China.
2. Materials and methods 2.1. Distribution data and phylogenetic treeWe used the same data set as Ye et al. (2019).
2.2. Distance metrics and clustering analysisTo compare the effect of richness-dependent and richnessindependent metrics on biogeographical regionalization, we used MNTD and pβsim to measure phylogenetic turnover (pβsim), respectively. MNTD is affected by differences in species richness whereas pβsim is independent of differences in species richness among grid cells (Slik et al., 2018). In this study, MNTD was obtained by the 'comdistnt' function of the 'picante' package (Kembel et al., 2010) in R (R Core Team, 2019) and pβsim was calculated according to Ye et al. (2019).
To identify spatial clusters, we compared the output from eight hierarchical clustering algorithms based on the MNTD and pβsim pairwise distance matrices:(1) single linkage (SL), (2) complete linkage (CL), (3) unweighted pair-group method using arithmetic averages (UPGMA), (4) unweighted pair-group method using centroids (UPGMC), (5) weighted pair-group method using arithmetic averages (WPGMA), (6) weighted pair-group method using centroids (WPGMC), and (7) and (8), two variants of Ward's minimum variance (ward.D and ward.D2). We used the Kelley-Gardner-Sutcliffe penalty function (Kelley et al., 1996) to get a reasonable number of clusters for the phytogeographical regions. The clustering algorithms and reasonable number of clusters selected for this study helped to identify appropriate phytogeographical regions, and show the differences of the floristic regions obtained by using MNTD and pβsim respectively.
2.3. Characterized clade and indicator genus analysesMNTD is sensitive to differences in taxon richness; therefore, we adopted floristic regionalization using pβsim. To identify clades that characterize a phytogeographical region (Ye et al., 2019) or contribute significantly to phylogenetic structure, we used the procedure nodesig in Phylocom 4.2 (Webb et al., 2008). The overall phylogenetic tree nodesig tests were used to further compare the genus pool of each phytogeographical region, and examine whether a node in the phylogeny has significantly more (sig. more) or significantly less (sig. less) descendent taxa in a sample than a null model predicts. The null model is a random draw of n taxa from the phylogeny, where n is the number of taxa in the sample (Webb et al., 2008).
We used the Dufrêne- Legendre indicator species analysis (Dufrêne and Legendre, 1997) to characterize the genus composition of each identified phytogeographical region. The indicator value (IndVal) was defined as the product of two quantities, specificity (A) and fidelity (B). A represents the probability that a surveyed site belongs to the target site group on the basis of a target genus that has been found (calculated as the relative frequency of the occurrence of each genus in the target site group, divided by the sum of relative frequencies over all groups). B is the probability of finding the target genus in a site that belongs to the site group (i.e., the relative frequency of a genus inside the target site group; De Câceres and Legendre, 2009). The significance of the IndVal was assessed by a permutation test (9, 999 permutations). A genus that had significant IndVals (P < 0.001) was ranked as a suitable indicator genus of its relevant phytogeographical region. The analysis was conducted using the 'indicspecies' package (De Câceres and Legendre, 2009).
3. Results 3.1. Floristic regionalization using MNTD and pβsimThe five clustering algorithms varied in performance for the MNTD pairwise distance analysis (Table 2). Ward's method was the best performing clustering algorithm (cophenetic coefficient = 0.99), followed by CL (complete linkage) (cophenetic coefficient = 0.97), while UPGMA (unweighted pair-group method using arithmetic averages) performed worst (cophenetic coefficient = 0.94). Therefore, we chose Ward's method. According to the Kelley-Gardner-Sutcliffe penalty function, eight floristic regions belonging to two groups were identified in China, with southern regions 6 and 8 forming one group (South Cluster), and the other six regions forming the other group (North Cluster) (Fig. 1a). The geographical dividing line between North and South Cluster in eastern China was roughly consistent with the Qinling Mountain-Huaihe River Line. The North Cluster further splits into two subclusters, with regions 1, 3 and 7 forming one subcluster and regions 2, 4 and 5 the other subcluster.
| Clustering algorithms | Cophenetic correlation coefficients |
| Unweighted pair-group method using arithmetic averages (UPGMA) | 0.94 |
| Weighted pair-group method using arithmetic averages (WPGMA) | 0.96 |
| Ward's method | 0.99 |
| Single (SL) | 0.95 |
| Complete linkage (CL) | 0.97 |
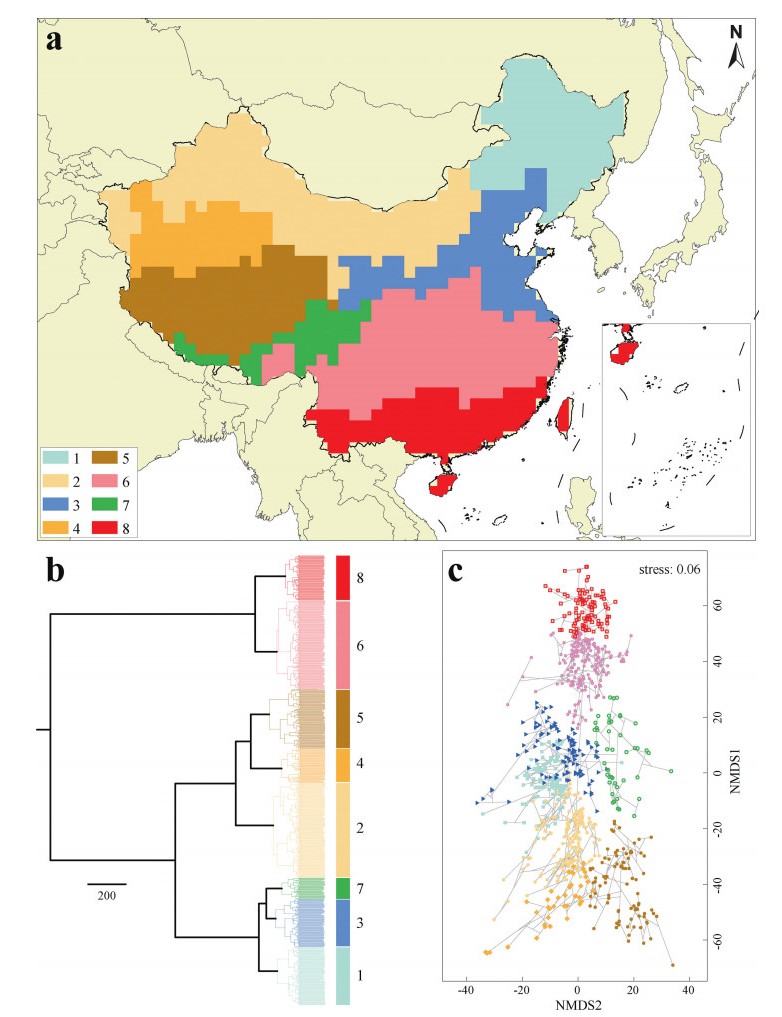
|
| Fig. 1 Map (a) and dendrogram (b) resulting from Ward hierarchical clustering and scatter plot from nonmetric multidimensional scaling (NMDS) two-dimensional ordination for the Chinese floristic assemblages based on MNTD distance matrices (c). The eight distinct floristic regions are highlighted in the dendrogram with large colored rectangles and displayed in the map in same colours. |
For the floristic regionalization using pβsim, five floristic regions were identified (Fig. 2; Fig. 4 in Ye et al., 2019).
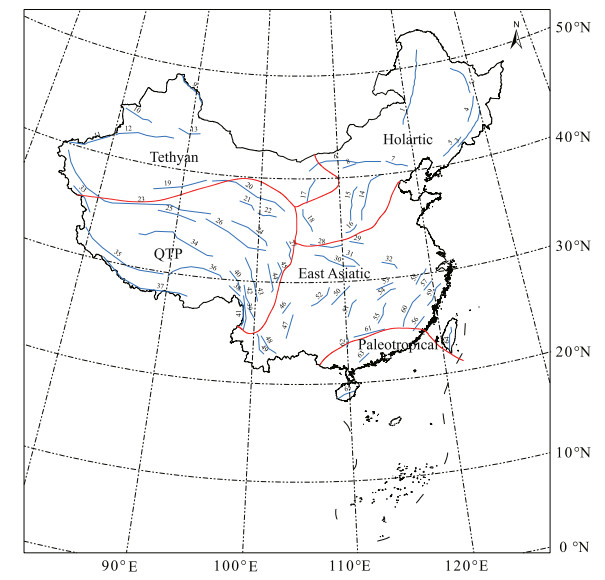
|
| Fig. 2 Map of the five floristic regions identified by pβsim and major mountain ranges in China. For the list of mountain ranges, see Table S6. The data for the elevation of China (a) and China's major mountain ranges (b) (Fig. 1a) were obtained from the National Fundamental Geographical Information System of China (http://nfgis.nsdi.gov.cn/nfgis/chinese/c_xz.htm). |
The nodesig algorithm identified differently characterized clades for different phytogeographical regions. The Tethyan region had the largest number (301 clades) of sig. more clades, which included 99.12% of genera (904 genera) in this region, but the mean number of genera contributed by each clade was the least (3.00 genera). In contrast, the East Asiatic region had relatively few sig. more clades (57 clades), which only included 42.88% of genera (988 genera) in this region, but each clade contributed the largest number (25.61 genera) of genera on average. Details of sig. more/sig. less nodes and the descendant taxa are shown in Appendix S1:Fig. S1.
Among the 2, 591 genera analyzed, 34.3% (889 genera) had a significant indicator value (Table 3). This was a substantial number of genera that qualified as indicator taxa and indicated that different phytogeographical regions had significantly different floras. The Paleotropic region had the largest proportion (40.6%, 361 genera) of total indicator genera (889 genera), whereas the Holarctic region had the least proportion of indicator genera (5.8%, 52 genera). The indicator genera identified for each phytogeographical region are provided in Appendix S1:Tables S1-S5.
| Phytogeographical region | Number of indictor genera (proportion) | Number of sig. more nodes | Number of sig. less nodes | Number of nodes | Number of genera descending from sig. more nodes | Number of genera | Mean number of genera descending from each sig. more nodes | The Proportion of regional genera descending from sig. more nodes |
| Paleotropic region | 342 (38.5%) | 158 | 271 | 2, 264 | 1, 340 | 1, 605 | 8.48 | 97.85% |
| Holarctic region | 52 (5.8%) | 152 | 184 | 2, 125 | 1, 227 | 1, 254 | 8.07 | 99.12% |
| East Asiatic region | 230 (25.9%) | 57 | 170 | 2, 539 | 1, 460 | 2, 304 | 25.61 | 42.88% |
| Tethyan region | 147 (16.5%) | 301 | 244 | 1, 661 | 904 | 912 | 3.00 | 63.37% |
| QTP region | 118 (13.3%) | 48 | 109 | 2, 472 | 813 | 1, 897 | 16.94 | 83.49% |
Beta diversity is composed of two different components:spatial species turnover and nestedness of assemblages, which result from two antithetic processes, namely species replacement and species loss, respectively (Lennon et al., 2001; Koleff et al., 2003; Baselga et al., 2007; Tuomisto, 2010). Using European longhorn beetles as a case, Baselga (2010) exemplified the relevance of disentangling spatial turnover and nestedness patterns. Baselga and Orme (2012) published an R package "betapart" to compute these dissimilarity measures. In the studies of biogeographical regionalization (bioregionalization), we should use the turnover component to measure composition dissimilarity between grid cells to avoid the impact of diversity differences (Leprieur and Oikonomou, 2014). Kreft and Jetz (2010) recommended the beta-sim index (βsim) and it has been applied in most of the subsequent bioregionalization studies (Table 1; Linder et al., 2012; lez-Orozco et al., 2013; Holt et al., 2013). However, some studies still used inappropriate or incorrect indices (e.g., Bray-Curtis, mean nearest phylogenetic distance), which have proved to be influenced by diversity differences (Slik et al., 2018). In this study, the floristic regions obtained by MNTD distance matrices were also influenced by diversity differences between grid cells, because the Chinese flora has a conspicuous east-west gradient in plant diversity (Lu et al., 2018).
4.2. Comparison between two regionalizationsThe cluster analysis based on MNTD distance matrices presented a picture of eight regions for the Chinese flora, whereas there were five regions based on pβsim identified by or found by Ye et al. (2019). The floristic regions obtained by MNTD distance matrices were similar with the patterns of generic richness in China (Extended Data Figure 8a in Lu et al., 2018). Southern regions 6 and 8 have the highest richness, regions 1, 3 and 7 have the moderate richness, and regions 2, 4 and 5 have the lowest richness. As a result, regionalization based on MNTD distance matrices was strongly influenced by the nestedness component of the metric (Baselga, 2012), which was also found in a study of Indo-Pacific coral reef fishes (Mouillot et al., 2013). Furthermore, the pβsim was demonstrated to be able to reflect the true phylogenetic turnover for the Chinese angiosperm flora (Fig. 2b in Ye et al., 2019) and the five floristic regions-the Paleotropic, Holarctic, East Asiatic, Tethyan, and QTP regions-which clearly reflected the evolutionary history of the flora (Ye et al., 2019).
In our study, the characterized clades identified by the nodesig algorithm for each phytogeographical region differed greatly. For example, the clades in magnoliids (Myristiaceae, Annonaceae, Magnoliaceae, Calycanthaceae, Lauraceae, Hernandiaceae, Piperaceae, and Aristolochiaceae) were significantly associated with the Paleotropic region, while clades in Asteraceae occurred less often than expected in this region but were overabundant in the Holarctic region (Appendix S1:Fig. S2). We found that in the East Asiatic region, the sig. more nodes were deeper nodes with the most descendant genera on average in the five regions. This suggests the East Asiatic region has older lineages in China, while the flora of the Tethyan region with shallower sig. more nodes is younger (Table 3).
We concluded the discussion with a description of the five regions (Paleotropic, Holarctic, East Asiatic, Tethyan, and QTP regions) identified by our analysis (Fig. 2).
4.3. The five identified regions based on the pβsim metricThe Paleotropic region consists of tropical evergreen broadleaved forest vegetation in the southeastern coast of China and Hainan Island. Its northern boundary is roughly the Tropic of Cancer (Fig. 2). Within the five regions, this region has the least amount of areas and lowest elevation, the highest mean annual temperature (MAT) and mean annual precipitation (MAP), and the richest indicator clades and genera (Table 1 in Ye et al., 2019, Table S1 in Appendix S1). The Paleotropic region has the most evolutionarily distinct flora in China (Table 1 in Ye et al., 2019) and is characterized by the presence of thermophilic Malesian taxa (e.g., Bruguiera, Chrysophyllum, Endospermum, Stylidium, Tetracera), which were immigrated from southern and southeastern Asia (Axelrod et al., 1998). These tropical and subtropical plant clades and genera have mostly not migrated to the subtropical and temperate areas in central and northern China and alpine areas in southwestern China, implying the prevalence of Niche Conservatism Hypothesis (Wiens and Donoghue, 2004), thus resulting in the main split of the Chinese flora between the southern Paleotropic and northern Chinese floras, as shown in our clustering dendrogram (Fig. 1b). The flora of the Paleotropic region is the oldest and represents a floristic museum (Lu et al., 2018).
The Holarctic region consists of Inner Mongolia steppe in the west and deciduous broad-leaved forest vegetation in Northeast and North China. Within North China, it is roughly bounded by the Helan Mountains in the west and the Taihang Mountains in the east (Fig. 2). The mean elevation is approximately 700 m, MAT is 4.1℃, and MAP is nearly 500 mm (Table 1 in Ye et al., 2019). Flora of this region includes 6, 878 species that belong to 1, 254 genera and 170 families. It represents the Boreal-Tertiary flora in China that preserved Northern Hemisphere temperate taxa with origins in the late Cretaceous and early Paleogene, such as Acer, Alnus, Corylus, Eleutherococcus, Rhododendron, and Tilia (Sun, 2002b). This region has the fewest indicator genera among the five regions that are mainly composed of north temperate taxa (e.g., Filifolium, Hypochaeris, Saposhnikovia, Scabiosa, and Trientalis) (Table 1 in Ye et al., 2019, Table S2 in Appendix S1).
The East Asiatic region includes deciduous broad-leaved forest vegetation in the east of the Taihang Mountains (Fig. 2), subtropical evergreen broad-leaved forest areas in most of southern China, and seasonal rain forest vegetation in southern Yunnan. Its western border extends along the western margin of the Sichuan Basin to the eastern and southern margin of the Yunnan Plateau. Flora of the East Asiatic region is closely related to that of the Holarctic region. The mean elevation is approximately 600 m a.s.l.; MAT is 16℃; MAP is nearly 1, 200 mm (Table 1 in Ye et al., 2019). The East Asiatic region is characterized by the presence of genera that are endemic to East Asia (e.g., Akebia, Emmenopterys, Pinellia) and genera with the distribution pattern of the East Asiae-North American disjunction patterns (e.g., Liriodendron, Shortia) (Table S3 in Appendix S1). The East Asiatic region hosts the highest plant species richness (20, 317 species) among the five regions (Table 3). First, as a stable and ancient area, the East Asiatic region preserved the flora from the early Cretaceous to early Paleogene (Wu et al., 2010). Second, with the uplift of the QTP, and intensification of both the Pacific and Indian Ocean monsoons in this area, new habitats have appeared during the Neogene, which might have promoted the rapid radiation of some plant taxa, such as Begoniaceae, Gesneriaceae, Orchidaceae, and Rubiaceae (Favre et al., 2015). Third, the East Asiatic region preserved Boreotropical plant taxa that were once common in the Northern Hemisphere but extinct from other areas during Quaternary glaciation, such as Cyclocarya, Davidia, Emmenopterys, and Eucommia (Wolfe, 1975; Wen, 1999; López-Pujol and Ren, 2010). Therefore, the East Asiatic region harbors both early and recently diverged taxa in China and East Asia.
The Tethyan region mainly includes the desert areas in North-west China (Fig. 2). The mean elevation is approximately 1, 800 m; MAT is 5.6℃; and MAP is 111.2 mm. The Tethyan region has the lowest MAP of the five regions. As a temperate desert flora, plant diversity in the Tethyan region is low, with only 5, 412 species that belong to 912 genera and 128 families (Table 1 in Ye et al., 2019). This region has 147 indicator genera, which are mainly temperate desert xerophytic shrubs and herbs (e.g., Ammopiptanthus, Glaucium, Lagochilus, Potaninia, Tugarinovia, and Zygophyllum) (Table S4 in Appendix S1). However, the Tethyan region had a high evolutionary distinctiveness in comparison with other regions (Table 1 in Ye et al., 2019). After the Oligocene, the inland Tethys Sea retreated and was accompanied by an increasingly dry climate, which resulted in diversification of the xerophytic Tethyan flora (Palamarev, 1989). Consequently, this region is the diversity center for Amaranthaceae (Chenopodioideae), Apiaceae, Brassicaceae, and Lamiaceae (Sun, 2013). Moreover, the Tethyan region was also the most important glacial refugium for Tertiary relics throughout Quaternary glacial cycles (Qiu et al., 2011); for example, Tetraena mongolica is considered a relic of the Tethys sea tropical phytogeographical region (Wu et al., 2010).
The QTP region is bordered to the south by the Himalayan Mountains, to the east by the Hengduan Mountains, to the north by the Kunlun Mountains and Qilian Mountains, and to the west by the Karakorum Mountains (Fig. 2). The QTP region is the largest region and has the highest average elevation (4, 248 m a.s.l.) and lowest MAT of the five regions. The QTP region is characterized by the presence of alpine genera (e.g., Cyananthus, Diapensia, Lancea, Meconopsis, Morina, and Solms-laubachia) (Table S5 in Appendix S1). This region possesses one of the world's richest flora, with 16, 220 species that belong to 1, 897 genera and 205 families (Table 1 in Ye et al., 2019), and numerous Chinese and local endemic species (Huang et al., 2016; Zhang et al., 2016a). Because of the uplift of the QTP, this region is also an important origin and diversification center of north temperate and alpine floras, such as the genera Aconitum, Corydalis, Gentiana, Rhododendron, Pedicularis, and Primula (Sun, 2002a; Zhang et al., 2016a).
5. ConclusionsWe demonstrated that the choice of metrics could have a dramatic impact on the biogeographical regionalization. Those regionalizations based on metrics that are sensitive to taxa richness can reflect the pattern of turnover and nestedness together, but cannot reflect the true pattern of taxonomic and phylogenetic turnover (Baselga, 2012). The regionalization based on the MNTD metric is greatly influenced by the richness differences in the Chinese flora. Our phylogeny-based regionalization using beta-sim metric provides a foundation for future biogeographical and biodiversity studies and can also be used to develop strategies to conserve taxa and phytogeographical regions with different origins and evolutionary histories more effectively in China.
Declaration of Competing InterestThe authors declare that there is no conflict of interest regarding this manuscript. All the authors agreed to submit this manuscript.
AcknowledgementsThis work was supported by grants from the National Natural Science Foundation of China (NNSF 31800178, 31590822, 31500179 and 31461123001), the Strategic Priority Research Program of Chinese Academy of Sciences (XDA19050103 and XDB31000000), the National Key Basic Research Program of China (2014CB954101).
Appendix A. Supplementary dataSupplementary data to this article can be found online at https://doi.org/10.1016/j.pld.2019.12.003.
Axelrod, D.I., Al-Shehbaz, I., Raven, P., 1998. In: Zhang, A., Wu, S. (Eds.), History of the modern flora of China. Floristic characteristics and diversity of East Asian plants. China Higher Education Press & Springer-Verlag, Beijing & Berlin, pp. 43-45.
|
Baselga A., 2010. Partitioning the turnover and nestedness components of beta diversity. Global Ecol. Biogeogr, 19: 134-143. DOI:10.1111/j.1466-8238.2009.00490.x |
Baselga A., 2012. The relationship between species replacement, dissimilarity derived from nestedness, and nestedness. Global Ecol. Biogeogr, 21: 1223-1232. DOI:10.1111/j.1466-8238.2011.00756.x |
Baselga A., Orme C.D.L., 2012. betapart: an R package for the study of beta diversity. Methods Ecol. Evol, 3: 808-812. DOI:10.1111/j.2041-210X.2012.00224.x |
Baselga A., Jim#233;nez-Valverde A., Niccolini G., 2007. A multiple-site similarity measure independent of richness. Biol. Lett, 3: 642-645. DOI:10.1098/rsbl.2007.0449 |
Bribiesca-Contreras G., Verbruggen H., Hugall A.F., O'Hara T.D., 2019. Global biogeographic structuring of tropical shallow-water brittle stars. J. Biogeogr, 46: 1287-1299. |
Clark D.R., Mathieu M., Mourot L., Dufoss#233; L., Underwood G.J.C., Dumbrell A.J., McGenity T.J., 2017. Biogeography at the limits of life: Do extremophilic microbial communities show biogeographical regionalization? Global Ecol. Biogeogr, 26: 1435-1446. |
Crisp M.D., Arroyo M.T.K., Cook L.G., Gandolfo M.A., Jordan G.J., McGlone M.S., Weston P.H., Westoby M., Wilf P., Linder H.P., 2009. Phylogenetic biome conservatism on a global scale.. Nature, 458: 754-756. DOI:10.1038/nature07764 |
Daru B.H., Michelle B., Olivier M., Kowiyou Y., Hanno S., Slingsby J.A., Jonathan D.T., 2016. A novel phylogenetic regionalization of phytogeographical zones of southern Africa reveals their hidden evolutionary affinities. J. Biogeogr, 43: 155-166. DOI:10.1111/jbi.12619 |
Daru B.H., Holt B.G., Lessard J.P., Yessoufou K., Davies T.J., 2017. Phylogenetic regionalization of marine plants reveals close evolutionary affinities among disjunct temperate assemblages. Biol. Conserv, 213: 351-356. DOI:10.1016/j.biocon.2016.08.022 |
De C#226;ceres M.D., Legendre P., 2009. Associations between species and groups of sites: indices and statistical inference. Ecology, 90: 3566-3574. DOI:10.1890/08-1823.1 |
Droissart V., Dauby G., Hardy O.J., et al, 2018. Beyond trees: biogeographical regionalization of tropical Africa. J. Biogeogr, 45: 1153-1167. DOI:10.1111/jbi.13190 |
Dufr#234;ne M., Legendre P., 1997. Species assemblages and indicator species: the need for a flexible asymmetrical approach. Ecol. Monogr, 67: 345-366. |
Favre A., Packert M., Pauls S.U., Jahnig S.C., Uhl D., Michalak I., MuellnerRiehl A.N., 2015. The role of the uplift of the Qinghai-Tibetan Plateau for the evolution of Tibetan biotas. Biol. Rev. Camb. Phil. Soc, 90: 236-253. DOI:10.1111/brv.12107 |
Godinho M.B.C., da Silva F.R., 2018. The influence of riverine barriers, climate, and topography on the biogeographic regionalization of Amazonian anurans. Sci.Rep, 8: 3427. DOI:10.1038/s41598-018-21879-9 |
Gonz#225;lez-Orozco C.E., Ebach M.C., Laffan S., Thornhill A.H., Knerr N.J., Schmidt-Lebuhn A.N., Cargill C.C., Clements M., Nagalingum N.S., Mishler B.D., 2014a. Quantifying phytogeographical regions of Australia using geospatial turnover in species composition. PloS One, 9: e92558-e92558. DOI:10.1371/journal.pone.0092558 |
Gonz#225;lez-Orozco C.E, Laffan S.W., Knerr N., Miller J.T., 2013. A biogeographical regionalization of Australian Acacia species. J. Biogeogr, 40: 2156-2166. DOI:10.1111/jbi.12153 |
Gonz#225;lez-Orozco C.E., Thornhill A.H., Knerr N., Laffan S., Miller J.T., Richardson D., 2014b. Biogeographical regions and phytogeography of the eucalypts. Divers. Distrib, 20: 46-58. DOI:10.1111/ddi.12129 |
Hattab T., Albouy C., Lasram F.B.R., Loc'H F.L., Guilhaumon F., Leprieur F., 2015. A biogeographical regionalization of coastal Mediterranean fishes. J. Biogeogr, 42: 1336-1348. DOI:10.1111/jbi.12505 |
He J., Kreft H., Gao E., Wang Z., Jiang H., 2017. Patterns and drivers of zoogeographical regions of terrestrial vertebrates in China. J. Biogeogr, 44: 1172-1184. DOI:10.1111/jbi.12892 |
Holt B.G., Jean-Philippe L., Borregaard M.K., Fritz S.A., Ara#250;jo M.B., Dimitar D., Pierre-Henri F., Graham C.H., Graves G.R., Jonsson K.A., 2013. An update of Wallace's zoogeographic regions of the world.. Science, 339: 74-79. DOI:10.1126/science.1228282 |
Huang J., Huang J., Liu C., Zhang J., Lu X., Ma K., 2016. Diversity hotspots and conservation gaps for the Chinese endemic seed flora. Biol. Conserv, 198: 104-112. DOI:10.1016/j.biocon.2016.04.007 |
J#248;nsson K.A., Holt B.G., 2015. Islands contribute disproportionately high amounts of evolutionary diversity in passerine birds. Nat. Commun, 6: 8538. DOI:10.1038/ncomms9538 |
Julien V., Andr#233; S. Ed V., Rogier V., Niels R., 2016. Phytogeography of New Guinean orchids: patterns of species richness and turnover. J. Biogeogr, 43: 204-214. DOI:10.1111/jbi.12612 |
Kelley L.A., Gardner S.P., Sutcliffe M.J., 1996. An automated approach for clustering an ensemble of NMR-derived protein structures into conformationally related subfamilies. Protein Eng, 9: 1063-1065. DOI:10.1093/protein/9.11.1063 |
Kembel S.W., Cowan P.D., Helmus M.R., Cornwell W.K., Morlon H., Ackerly D.D., Blomberg S.P., Webb C.O., 2010. Picante: R tools for integrating phylogenies and ecology.. Bioinformatics, 26: 1463-1464. DOI:10.1093/bioinformatics/btq166 |
Koleff P., Gaston K.J., Lennon J.J., 2003. Measuring beta diversity for presenceeabsence data. J. Anim. Ecol, 72: 367-382. DOI:10.1046/j.1365-2656.2003.00710.x |
K#246;nig C., Weigelt P., Kreft H., 2017. Dissecting global turnover in vascular plants. Global Ecol. Biogeogr, 26: 228-242. DOI:10.1111/geb.12536 |
Kreft H., Jetz W., 2010. A framework for delineating biogeographical regions based on species distributions. J. Biogeogr, 37: 2029-2053. DOI:10.1111/j.1365-2699.2010.02375.x |
Kubota Y., Hirao T., Fujii S.-J., Shiono T., Kusumoto B., 2014. Beta diversity of woody plants in the Japanese archipelago: the roles of geohistorical and ecological processes. J. Biogeogr, 41: 1267-1276. DOI:10.1111/jbi.12290 |
Lennon J.J., Koleff P., Greenwood J.J.D., Gaston K.J., 2001. The geographical structure of British bird distributions: diversity, spatial turnover and scale. J. Anim. Ecol, 70: 966-979. |
Leprieur F., Oikonomou A., 2014. The need for richness-independent measures of turnover when delineating biogeographical regions. J. Biogeogr, 41: 417-420. DOI:10.1111/jbi.12266 |
Li R., Kraft N.J.B., Yang J., Wang Y., 2015. A phylogenetically informed delineation of floristic regions within a biodiversity hotspot in Yunnan, China.. Sci. Rep, 5: 9396. DOI:10.1038/srep09396 |
Li R., Qian L., Sun H., 2018. Current progress and future prospects in phylofloristics. Plant Divers, 40: 141-146. DOI:10.1016/j.pld.2018.07.003 |
Li R., Sun H., 2017. Phylofloristics: a case study from Yunnan, China. Biodivers. Sci, 25: 195-203. |
Linder H.P., Lovett J.C., Mutke J., Barthlott W., J#252;rgens N., Rebelo T., Küper W., 2005. A numerical re-evaluation of the sub-Saharan phytochoria of mainland Africa. Biol. Skr, 55: 229-252. |
Linder H.P., Klerk H.M.D., Born J., Burgess N.D., Fjelds#229; J., Rahbek C., 2012. The partitioning of Africa: statistically defined biogeographical regions in subSaharan Africa. J. Biogeogr, 39: 1189-1205. DOI:10.1111/j.1365-2699.2012.02728.x |
López-Pujol, J., Ren, M., 2010. China: a hot spot of relict plant taxa. In: Rescigno, V., Maletta, S. (Eds.), Biodiversity Hotspots. Nova Science Publishers, New York, pp. 123-137.
|
Lu L.M., Mao L.F., Yang T., Ye J.F., Liu B., Li H.L., Sun M., et al, 2018. Evolutionary history of the angiosperm flora of China.. Nature, 554: 234-238. DOI:10.1038/nature25485 |
Mackey B., Berry S.L., Brown T., 2007. Reconciling approaches to biogeographical regionalization: a systematic and generic framework examined with a case study of the Australian continent. J. Biogeogr, 35: 213-229. DOI:10.1111/j.1365-2699.2007.01822.x |
Meyer R.S., DuVal A.E., Jensen H.R., 2012a. Patterns and processes in crop domestication: an historical review and quantitative analysis of 203 global food crops. New Phytol, 196: 29-48. DOI:10.1111/j.1469-8137.2012.04253.x |
Meyer R.S., Karol K.G., Little D.P., Nee M.H., Litt A., 2012b. Phylogeographic relationships among Asian eggplants and new perspectives on eggplant domestication. Mol. Phylogenet. Evol, 63: 685-701. DOI:10.1016/j.ympev.2012.02.006 |
Mouillot D., De Bortoli J., Leprieur F., Parravicini V., Kulbicky M., Bellwood D.R., 2013. The challenge of delineating biogeographical regions: nestedness matters for Indo-Pacific coral reef fishes. J. Biogeogr, 40: 2228-2237. DOI:10.1111/jbi.12194 |
Palamarev E., 1989. Paleobotanical evidences of the Tertiary history and origin of the Mediterranean sclerophyll dendroflora. Plant Systemat. Evol, 162: 93-107. DOI:10.1007/BF00936912 |
Qiu Y.X., Fu C.X., Comes H.P., 2011. Plant molecular phylogeography in China and adjacent regions: tracing the genetic imprints of Quaternary climate and environmental change in the world's most diverse temperate flora. Mol. Phylogenet. Evol, 59: 225-244. DOI:10.1016/j.ympev.2011.01.012 |
R Core Team, 2019. R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria.
|
Rueda M., Rodríguez M.Á., Hawkins B.A., 2013. Identifying global zoogeographical regions: lessons from Wallace. J. Biogeogr, 40: 2215-2225. DOI:10.1111/jbi.12214 |
Silva A.C., Souza A.F., 2018. Aridity drives plant biogeographical sub regions in the Caatinga, the largest tropical dry forest and woodland block in South America.. PloS One, 13: e0196130. DOI:10.1371/journal.pone.0196130 |
Slik J.W.F., Franklin J., Arroyo-Rodr#237;guez V., Field R., Aguilar S., Aguirre N., Ahumada J., Aiba S.I., Alves L.F., Anitha K., Avella A., Mora F., et al, 2018. Phylogenetic classification of the world's tropical forests. Proc. Natl. Acad. Sci. U. S. A, 115: 1837-1842. DOI:10.1073/pnas.1714977115 |
Sun H., 2002a. Tethys retreat and Himalayas-Hengduanshan Mountains uplift and their significance on the origin and development of the Sino-Himalayas elements and alpine flora.. Acta Bot. Yunnanica, 24: 273-288. |
Sun H., 2002b. Evolution of arctic tertiary flora in Himalayan-Hengduan Mountains.. Acta Bot. Yunnanica, 24: 671-688. |
Sun, H., 2013. In: Hong, D.Y., Blackmore, S. (Eds.), Phytogeographical regions of China, Plants of China. Science Press, Beijing, pp. 176-204.
|
Takhtajan, A., 1978. Floristic regions of the world. Academy of Sciences of the U.S.S.R., Leningrad.
|
Tuomisto H., 2010. A diversity of beta diversities: straightening up a concept gone awry. Part 1. Defining beta diversity as a function of alpha and gamma diversity.. Ecography, 33: 2-22. DOI:10.1111/j.1600-0587.2009.05880.x |
Webb C.O., 2000. Exploring the phylogenetic structure of ecological communities:an example for rain forest trees. Am. Nat, 156: 145-155. DOI:10.1086/303378 |
Webb C.O., Ackerly D.D., Kembel S.W., 2008. Phylocom: software for the analysis of phylogenetic community structure and trait evolution.. Bioinformatics, 24: 2098-2100. DOI:10.1093/bioinformatics/btn358 |
Wen J., 1999. Evolution of eastern Asian and eastern North American disjunct distributions in flowering plants. Annu. Rev. Ecol. Systemat, 30: 421-455. |
Wiens J.J., Donoghue M.J., 2004. Historical biogeography, ecology and species richness. Trends Ecol. Evol, 19: 639-644. DOI:10.1016/j.tree.2004.09.011 |
Wolfe J.A., 1975. Some aspects of plant geography of the northern hemisphere during the late Cretaceous and Tertiary. Ann. Mo. Bot. Gard, 62: 264-279. DOI:10.2307/2395198 |
Wu, Z.Y., Sun, H., Zhou, Z.K., Li, D.Z., Peng, H., 2010. Floristics of seed plants from China. Science Press, Beijing (in Chinese).
|
Xie Y., Mackinnon J., Li D., 2004. Study on biogeographical divisions of China. Biodivers. Conserv, 13: 1391-1417. DOI:10.1023/B:BIOC.0000019396.31168.ba |
Ye J.F., Lu L.M., Liu B., Yang T., Zhang J.L., Hu H.H., Li R., Lu A.M., Liu H.Y., Mao L.F., Chen Z.D., 2019. Phylogenetic delineation of regional biota: A case study of the Chinese flora. Mol. Phylogenet. Evol, 135: 222-229. |
Zhang D., Ye J., Sun H., 2016a. Quantitative approaches to identify floristic units and centres of species endemism in the Qinghai-Tibetan Plateau, south-western China. J. Biogeogr, 43: 2465-2476. DOI:10.1111/jbi.12819 |
Zhang M., Slik J.W.F., Ma K., 2016b. Using species distribution modeling to delineate the botanical richness patterns and phytogeographical regions of China. Sci. Rep, 6: 22400. DOI:10.1038/srep22400 |



