b. Plant Germplasm and Genomics Center, The Germplasm Bank of Wild Species, Kunming Institute of Botany, Chinese Academy of Sciences, Kunming 650201, China;
c. Institute of Tibetan Plateau Research at Kunming, Kunming Institute of Botany, Chinese Academy of Sciences, Kunming 650201, China;
d. University of the Chinese Academy of Sciences, Beijing 100049, China
Genes of the Lateral Organ Boundaries Domain/ASYMMETRIC LEAVES2-like (LBD/AS2) protein family encode plant-specific transcription factors that share a highly conserved LOB domain (Iwakawa et al., 2002; Xu et al., 2016). The conserved lateral organ boundaries domain (LBD) consists of 100 amino acids and contains a conserved CX2CX6CX3C zinc finger-like block for DNA-binding activity, a leucine-zipper-like coiled-coil motif for protein-protein interaction and a GAS (Gly-Ala-Ser) block which may also play a role in DNA-binding (Lee et al., 2013; Majer and Hochholdinger, 2011; Shuai et al., 2002; Xu et al., 2016). The variable C-terminal region of the LBD proteins plays a critical role in controlling the expression of downstream target genes (Liu et al., 2005; Majer et al., 2012). LBD proteins have been divided into two clades (class Ⅰ and class Ⅱ) according to their structure. In various species, most members of class Ⅰ have a complete LOB domain and all members of class Ⅱ have an incomplete leucine-zipper-like coiled-coil motif (Cao et al., 2016; Lu et al., 2017; Majer et al., 2012; Wang et al., 2013; Yang et al., 2006; Yordanov et al., 2010). In barley, mulberry, rice, grape, Arabidopsis, maize and apple, the number of LBD gene family members varies from 24 to 58 (Cao et al., 2016; Guo et al., 2016; Luo et al., 2016; Shuai et al., 2002; Wang et al., 2013; Yang et al., 2006; Zhang et al., 2014). The spatio-temporal expression profiles of LBD genes vary, implying that during growth and development LBD proteins function at specific periods and in specific organs (Cao et al., 2016; Shuai et al., 2002; Wang et al., 2013).
Previous studies have demonstrated that LBD proteins play important roles in plant growth, development, signal transduction, and stress response development (Cabrera et al., 2014; Chen et al., 2012; Cortizo and Laufs, 2012; Fan et al., 2012; Feng et al., 2012; Hu et al., 2014; Kim et al., 2015; Lee et al., 2017; Liu et al., 2014; Mangeon et al., 2011; Oh et al., 2010; Okushima et al., 2007; Thatcher et al., 2012; Yordanov et al., 2010; Zhou et al., 2012). For example, Arabidopsis LBD proteins (AtLBD16, AtLBD18, AtLBD29 and AtLBD33) have been shown to act in the auxin-mediated signaling pathway that regulates lateral root development (Okushima et al., 2007; Xu et al., 2016). More recently, research has demonstrated that the dimerization in transcription factors LBD16 and LBD18 plays a crucial role for lateral root formation (Lee et al., 2017). Furthermore, transcription factors from class Ⅱ LBD genes (AtLBD37, AtLBD38 and AtLBD39) have been shown to repress the biosynthesis of anthocyanin and regulate the nitrogen metabolism (Rubin et al., 2009).
Turnip (Brassica rapa var. rapa) is an important vegetable crop in the Tibetan Plateau. However, no report on the turnip LBD gene family exists. The turnip genome has been sequenced and assembled (Lin et al., 2014), providing the basis for characterizing the LBD gene family in turnip.
In this study, 59 BrrLBD genes were identified in the turnip genome, their phylogenetic relationship, exon/intron structure, protein motifs and chromosome locations were analyzed. Furthermore, we characterized the expression profiles of BrrLBD genes in different tissues.
Material and methods Plant material and growth conditionsB. rapa var. rapa 'KTRG-B36' from Lanping county, Nujiang City, Yunnan Province, China, was used. For root collection, seedlings were grown on a Whatman filter paper and watered with 1/2 MS medium for one week. For other tissues, plants were grown in a greenhouse (22 ℃) under long-day conditions (16 h light/day). Leaves and shoot apical meristem were derived from 8-week-old plants; floral buds were derived from 10-week-old plants.
Identification of turnip LBD family genesThe whole genome sequence of B. rapa var. rapa (Lin et al., 2014) was downloaded from www.bioinformatics.nl/brassica/turnip. Forty-two Arabidopsis LBD genes were downloaded from TAIR 12.0 (www.arabidopsis.org). All 43 known Arabidopsis LBDs were used as queries to search the turnip genome database at an e-value cutoff of 1e-003 (Du et al., 2017; Lu et al., 2017). The pI (isoelectric points), MW (protein molecular weights) and GRAVY (Grand average of hydropathicity) were obtained by proteomic and sequence analysis tools of the ExPASy proteomics server (http://expasy.org/) (Artimo et al., 2012). Chromosomal locations were found in the turnip database by using a Perl-based program.
Gene structure and conserved motif analysesBrrLBD gene exon and intron structures were identified with Gene Structure Display Server 2.0 (GSDS, http://gsds.cbi.pku.edu.cn/) (Hu et al., 2015). Conserved motifs of the BrrLBD proteins were analyzed using the Multiple Em for Motif Elucidation (MEME) program (http://meme-suite.org/index.html) (Bailey and Elkan, 1994) with the following parameters: (1) the optimum motif width was set from 6 to 200, and (2) the maximum number of motifs was set to identify 20 motifs.
Phylogenetic analysisAll phylogenetic trees of protein sequences were constructed with the neighbor-joining method using the MEGA 7.0 program. The reliability of the trees was tested by the bootstrapping method with 1000 replicates. All images of phylogenetic trees were drawn in MEGA 7.0 (Kumar et al., 2016).
Chromosome distribution, and divergence time estimation of BrrLBD genesBrrLBD genes were mapped on chromosomes by confirming their detailed chromosomal positions supplied by the Turnip Genome Database. To determine their physical location, the starting positions of all BrrLBD genes on each chromosome were confirmed based on a local database of the complete sequence of the turnip genome by searching BLASTn. Segmental and tandem duplication regions were obtained from MCscanX. Analysis of synteny blocks in turnip genes was visualized using Circos (http://circos.ca/). Synonymous (Ks) and nonsynonymous (Ka) substitution rates were estimated using the codeml program of the PAML4 package (Yang, 2007). The divergence time (T) of turnip gene pairs was calculated using formula T = Ks/2λ, where λ represents the divergence rate of 1.5 × 10-8 for Arabidopsis (Gaut et al., 1996).
Quantitative RT-PCRTotal RNA was extracted from KTRG-B36 leaves and shoot apical meristem of 8-week-old plants, floral buds of 10-week-old plants using EastepTM Universal RNA Extraction Kit (Promega, Shanghai, China). All quantitative real-time PCR primers were designed by the online NCBI program Primer-BLAST (https://www.ncbi.nlm.nih.gov/tools/primer-blast/) applying the following parameters: 150-200 bp of polymerase chain reaction (PCR) product size, 58 ℃-62 ℃ primer melting temperatures (Tm), and organism (taxid:3711) for B. rapa. At least one of two primers was derived from differential sequences of highly similar genes. qRT-PCR was performed in three technical repetitions with cDNAs synthesized from three biological replicates of KTRG-B36 different tissues. BrrTUB2 was used as reference gene.
Subcellular localization and confocal laser scanning microscopyThe coding region sequence (CDS) of BrrLBD genes were subcloned into pRI101-GFP using the EZ-cloning method. 35S:GFP-BrrLBDs were transferred into Agrobacterium tumefaciens EHA105 using electroporation. Positive transformants were prepared for leaf infiltration according to Du et al. (2017), and laser confocal microscopy was performed.
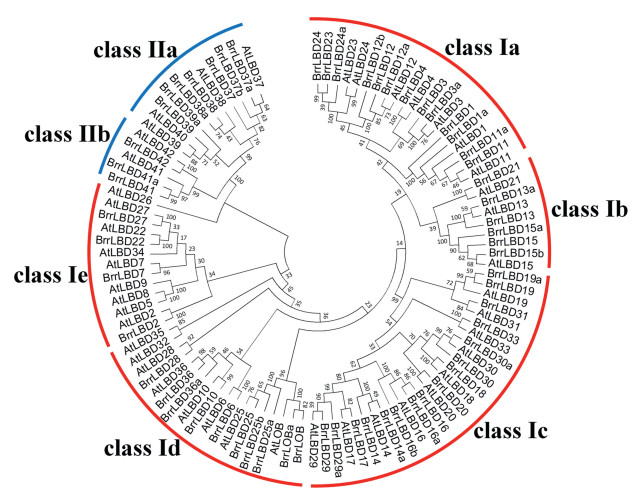
Results Identification and annotation of LBD genes in turnipsThe release of the complete turnip genome sequences allowed the genome-wide identification of turnip genes (Lin et al., 2014). In the present study, BLAST was used to search BrrLBDs in the turnip genome. The obtained sequences were further verified through the Hidden Markov Model of the SMART and pfam 31.0. A total of 59 LBD-like sequences were identified from the turnip genome, all of which possessed conserved LOB motifs. The BrrLBD genes were annotated following the nomenclature of Arabidopsis thaliana depending on protein sequence similarities (Fig. 1).
|
| Fig. 1 Phylogenetic tree of LBD proteins from turnip and Arabidopsis The evolutionary history of LBD proteins was inferred by using 59 BrrLBD protein and 43 AtLBD protein sequences to construct a Neighbor-Joining (NJ) cluster tree. The percentage of replicate trees in which the associated taxa clustered together in the bootstrap test (1000 replicates) is shown next to the branches. Evolutionary analyses were conducted in MEGA7. |
BrrLBD proteins were classified into seven subclades (subclades a to e within class Ⅰ, and subclades a and b within class Ⅱ). Among them, most BrrLBDs were clustered into class Ⅰ. Thirteen BrrLBDs were classified as class Ⅰa; 6 BrrLBDs fell into class Ⅰb; 16 BrrLBDs were included in class Ⅰc, 10 BrrLBDs were clustered into class Ⅰd, 4 BrrLBDs were classified as class Ⅰe. Sequence analysis revealed that AtLBD5, 8, 9, 26, 32, 34 and 36 had no orthologs in turnip. AtLOB, AtLBD3, 11, 12, 13, 14, 15, 16, 19, 24, 25, 29, 30, 37, 38, 39 and 41 had more than one ortholog in the turnip genome. The standard annotations of 59 BrrLBD genes are shown in Table 1; accession numbers for sequences were submitted to GenBank.
| Gene Name | Accession Numbe | CDS length (bp) | Protein size (aa) | MV(kD) | PI | GRAVY | Chr. Location |
| BrrLOB | MF953601 | 546 | 181 | 19.91 | 8.29 | -0.442 | Chr06:14089874..14090419 |
| BrrLOBa | MF953602 | 531 | 176 | 19.27 | 8.56 | -0.453 | Chr09:3249530..3250060 |
| BrrLBD1 | MF953603 | 624 | 207 | 23.13 | 4.87 | -0.267 | Chr09:35566092..35567209 |
| BrrLBD1a | MF953604 | 723 | 240 | 26.61 | 5.36 | -0.215 | Chr06:2669916..2671082 |
| BrrLBD2 | MF953605 | 594 | 197 | 22.49 | 9.68 | -0.909 | Chr08:20861229..20861822 |
| BrrLBD3 | MF953606 | 492 | 163 | 18.08 | 8.53 | -0.187 | Chr06:6348336..6349858 |
| BrrLBD3a | MF953607 | 495 | 164 | 18.32 | 9.22 | -0.200 | Chr09:33285226..33286439 |
| BrrLBD4 | MF953608 | 519 | 172 | 18.67 | 7.61 | -0.361 | Chr07:5298495..5299499 |
| BrrLBD6 | MF953609 | 609 | 202 | 22.09 | 7.10 | -0.375 | Chr02:9571227..9571835 |
| BrrLBD7 | MF953610 | 558 | 185 | 21.10 | 8.30 | -0.350 | Chr02:12903480..12904037 |
| BrrLBD10 | MF953611 | 927 | 308 | 34.41 | 4.83 | -0.640 | Chr04:10470622..10471548 |
| BrrLBD11 | MF953612 | 699 | 232 | 25.53 | 4.92 | -0.308 | Chr04:12704785..12706200 |
| BrrLBD11a | MF953613 | 696 | 231 | 25.40 | 4.90 | -0.376 | Chr03:11340842..11342322 |
| BrrLBD12 | MF953614 | 573 | 190 | 21.13 | 5.91 | -0.354 | Chr04:13390300..13391118 |
| BrrLBD12a | MF953615 | 570 | 189 | 21.08 | 6.03 | -0.320 | Chr05:7475733..7476562 |
| BrrLBD12b | MF953616 | 750 | 249 | 28.02 | 7.09 | -0.368 | Chr03:6995346..6998651 |
| BrrLBD13 | MF953617 | 807 | 268 | 29.28 | 8.98 | -0.523 | Chr05:7375325..7377428 |
| BrrLBD13a | MF953618 | 801 | 266 | 28.87 | 9.04 | -0.411 | Chr04:13502689..13505830 |
| BrrLBD14 | MF953619 | 543 | 180 | 20.20 | 5.48 | -0.216 | Chr05:6926370..6927053 |
| BrrLBD14a | MF953620 | 543 | 180 | 20.27 | 5.00 | -0.248 | Chr04:14150149..14150839 |
| BrrLBD15 | MF953621 | 681 | 226 | 24.79 | 7.69 | -0.419 | Chr04:17194524..17195851 |
| BrrLBD15a | MF953622 | 672 | 223 | 24.46 | 8.73 | -0.432 | Chr05:830241..831235 |
| BrrLBD15b | MF953623 | 579 | 192 | 20.67 | 8.23 | -0.255 | Chr03:9761557..9762501 |
| BrrLBD16 | MF953624 | 741 | 246 | 26.62 | 8.11 | -0.468 | Chr04:17743892..17745306 |
| BrrLBD16a | MF953625 | 735 | 244 | 26.27 | 8.15 | -0.403 | Chr05:1354391..1355425 |
| BrrLBD16b | MF953626 | 717 | 238 | 25.95 | 6.88 | -0.372 | Chr03:10167241..10169052 |
| BrrLBD17 | MF953627 | 600 | 199 | 22.59 | 6.23 | -0.524 | Chr04:17751076..17751838 |
| BrrLBD18 | MF953628 | 780 | 259 | 27.10 | 8.51 | -0.224 | Chr04:18509449..18511269 |
| BrrLBD19 | MF953629 | 567 | 188 | 20.74 | 6.29 | -0.256 | Chr05:2384192..2384849 |
| BrrLBD19a | MF953630 | 507 | 168 | 18.60 | 7.68 | -0.179 | Chr04:18502804..18503399 |
| BrrLBD20 | MF953631 | 828 | 275 | 29.14 | 7.27 | -0.300 | Chr07:496401..497739 |
| BrrLBD21 | MF953632 | 618 | 205 | 22.77 | 8.28 | -0.135 | Chr05:21715551..21716168 |
| BrrLBD22 | MF953633 | 774 | 257 | 29.51 | 5.62 | -0.755 | Chr05:20593305..20594078 |
| BrrLBD23 | MF953634 | 357 | 118 | 13.31 | 9.17 | -0.305 | Chr03:16849276..16849721 |
| BrrLBD24 | MF953635 | 366 | 121 | 13.59 | 8.90 | -0.267 | Chr06:22637883..22638342 |
| BrrLBD24a | MF953636 | 366 | 121 | 13.84 | 9.03 | -0.347 | Chr02:22273191..22273670 |
| BrrLBD25 | MF953637 | 396 | 131 | 14.58 | 7.60 | -0.344 | Chr09:1251553..1251948 |
| BrrLBD25a | MF953638 | 399 | 132 | 14.56 | 7.58 | -0.314 | Chr06:22268459..22268857 |
| BrrLBD25b | MF953639 | 399 | 132 | 14.68 | 6.89 | -0.391 | Chr02:21758022..21758420 |
| BrrLBD27 | MF953640 | 1041 | 346 | 39.36 | 5.15 | -0.605 | Chr06:10206064..10207198 |
| BrrLBD28 | MF953641 | 537 | 178 | 20.08 | 6.58 | -0.553 | Chr01:15707079..15707615 |
| BrrLBD29 | MF953642 | 678 | 225 | 24.80 | 6.21 | -0.478 | Chr09:28873054..28873853 |
| BrrLBD29a | MF953643 | 660 | 219 | 24.32 | 6.28 | -0.481 | Chr04:1562942..1563718 |
| BrrLBD30 | MF953644 | 675 | 224 | 23.73 | 6.53 | -0.164 | Chr02:16017746..16020434 |
| BrrLBD30a | MF953645 | 675 | 224 | 23.59 | 6.69 | -0.056 | Chr09:1062280..1064275 |
| BrrLBD31 | MF953646 | 669 | 222 | 24.23 | 6.16 | -0.378 | Chr09:1068061..1069446 |
| BrrLBD33 | MF953647 | 552 | 183 | 20.62 | 5.72 | -0.183 | Chr10:15419817..15420448 |
| BrrLBD36 | MF953648 | 1011 | 336 | 37.36 | 6.56 | -0.645 | Chr07:9891804..9892814 |
| BrrLBD36a | MF953649 | 1185 | 394 | 44.24 | 5.34 | -0.726 | Chr09:4268544..4269728 |
| BrrLBD37 | MF953650 | 711 | 236 | 25.72 | 9.03 | -0.333 | Chr09:3854563..3855367 |
| BrrLBD37a | MF953651 | 726 | 241 | 25.99 | 8.46 | -0.337 | Chr02:27506908..27507701 |
| BrrLBD37b | MF953652 | 771 | 256 | 27.87 | 8.40 | -0.398 | Chr07:9589107..9589970 |
| BrrLBD38 | MF953653 | 738 | 245 | 26.70 | 6.82 | -0.249 | Chr09:24819925..24820971 |
| BrrLBD38a | MF953654 | 711 | 236 | 25.99 | 6.34 | -0.320 | Chr03:21473516..21474319 |
| BrrLBD39 | MF953655 | 714 | 237 | 26.02 | 7.55 | -0.312 | Chr01:627066..627939 |
| BrrLBD39a | MF953656 | 699 | 232 | 25.30 | 9.10 | -0.119 | Chr03:30663003..30663878 |
| BrrLBD41 | MF953657 | 795 | 264 | 28.18 | 8.77 | -0.308 | Chr01:25183419..25184297 |
| BrrLBD41a | MF953658 | 780 | 259 | 27.76 | 8.51 | -0.214 | Chr03:14495479..14496347 |
| BrrLBD42 | MF953659 | 714 | 237 | 25.81 | 8.05 | -0.355 | Chr07:17868701..17869608 |
| MW, molecular weight; PI, isoelectric point; GRAVY, Grand average of hydropathicity. | |||||||
The BrrLBD proteins were predicted to encode polypeptides of 118-394 amino acids with molecular weights ranging from 13.31 to 44.24 kDa. The theoretical pI ranged from 4.83 to 9.68. All values of GRAVY were less than zero, indicating that all polypeptides of BrrLBD protein are hydrophilic.
BrrLBD genes in turnip are distributed in all 10 turnip chromosomes (Fig. 2). The maximum number of BrrLBD genes per chromosome was found for chromosome A04 with 11 BrrLBD genes. Ten genes were found at chromosome 9, eight genes each were located at chromosome 3 and 5, and six genes each were located at chromosome 2 and 6, respectively. Chromosome 8 and 10 contain the minimum numbers of BrrLBD genes, with only one each. Turnip chromosome 7 has five LBD genes, and the chromosome 1 contains three BrrLBD genes.

|
| Fig. 2 Chromosome distribution of BrrLBD genes Turnip Chromosomes are shown in different colors in the outer circle, where the numbers represent the chromosome length in 100 Kb. The BrrLBD genes are marked at the approximate positions with specific color lines on the circle. Filled blocks in different colors represent the syntenic relationships of BrrLBD genes. |
In this study, a total of 17 pairs of putative LBD paralog proteins were found in the segmental duplication blocks distributed in different chromosomes. These results demonstrate that the expansion of BrrLBD gene family in turnip was involved in largescale segmental duplication events.
The divergence time (T) of seventeen pairs of BrrLBD paralog proteins was estimated by measuring the synonymous (Ks) and nonsynonymous (Ka) mutation rates (Table 2). The estimated divergence time (T) for BrrLBD duplicated gene pairs was approximately from 3.9 to 24.30 million years ago (MYA), with an average duplication time of ~10.4 MYA. The ω-values (Ka/Ks) for the BrrLBD paralogs were less than one. However, the duplicated pair BrrLBD19/BrrLBD19a (ω = 0.5624) had a relatively large ω value, which implies that this pair may have evolved rapidly from the last common ancestor.
| Seq1 | Seq2 | Identity (%) | Ks | Ka | ω | T (MYA) |
| BrrLBD12 | BrrLBD12a | 0.9319 | 0.1616 | 0.0202 | 0.1250 | 5.3867 |
| BrrLBD23 | BrrLBD24 | 0.9008 | 0.1784 | 0.0536 | 0.3004 | 5.9467 |
| BrrLBD3 | BrrLBD3a | 0.8354 | 0.3331 | 0.0585 | 0.1756 | 11.1033 |
| BrrLBD11 | BrrLBD11a | 0.8397 | 0.3155 | 0.0245 | 0.0777 | 10.5167 |
| BrrLOB | BrrLOBa | 0.8798 | 0.2346 | 0.0205 | 0.0874 | 7.8200 |
| BrrLBD25 | BrrLBD25a | 0.9167 | 0.4132 | 0.0334 | 0.0808 | 13.7733 |
| BrrLBD13 | BrrLBD13a | 0.8963 | 0.3768 | 0.0102 | 0.0271 | 12.5600 |
| BrrLBD15 | BrrLBD15b | 0.7832 | 0.3670 | 0.0288 | 0.0785 | 12.2333 |
| BrrLBD19 | BrrLBD19a | 0.6974 | 0.1161 | 0.0653 | 0.5624 | 3.8700 |
| BrrLBD30 | BrrLBD30a | 0.8705 | 0.3602 | 0.0332 | 0.0922 | 12.0067 |
| BrrLBD16 | BrrLBD16a | 0.9024 | 0.2881 | 0.0081 | 0.0281 | 9.6033 |
| BrrLBD14 | BrrLBD14a | 0.8556 | 0.2311 | 0.0535 | 0.2315 | 7.7033 |
| BrrLBD29 | BrrLBD29a | 0.8451 | 0.2743 | 0.0323 | 0.1178 | 9.1433 |
| BrrLBD41 | BrrLBD41a | 0.8491 | 0.2845 | 0.0041 | 0.0144 | 9.4833 |
| BrrLBD39 | BrrLBD39a | 0.7890 | 0.2530 | 0.0484 | 0.1913 | 8.4333 |
| BrrLBD38 | BrrLBD38a | 0.8462 | 0.7290 | 0.0165 | 0.0226 | 24.3000 |
| BrrLBD37 | BrrLBD37a | 0.8272 | 0.3750 | 0.0206 | 0.0549 | 12.5000 |
| Ks, synonymous substitution rates; Ka, Nonsynonymous substitution rates; MYA, million years ago. | ||||||
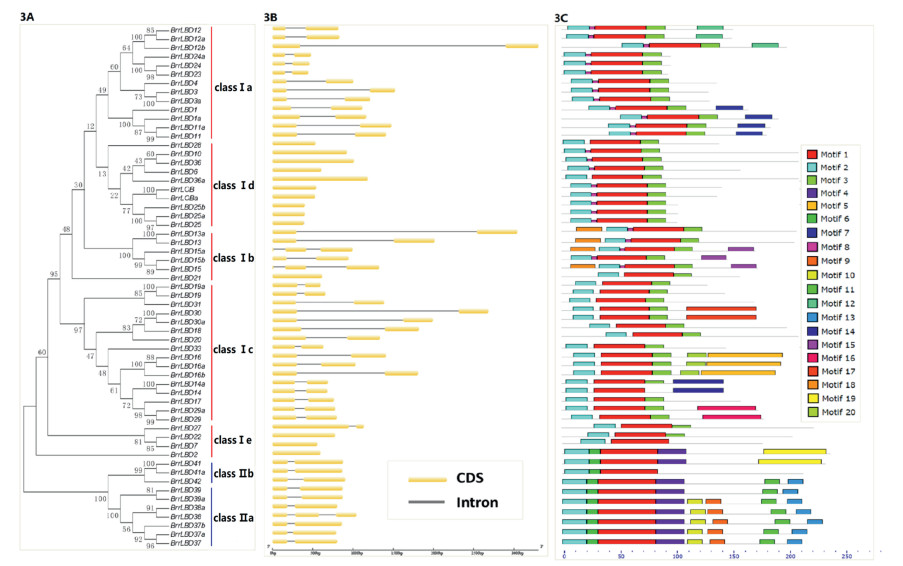
To mine information concerning exon/intron organization of BrrLBDs and conserved motifs of BrrLBDs, a new neighbor-joining (NJ) tree was constructed using the protein sequences of BrrLBDs (Fig. 3A). Gene structure analysis revealed that most BrrLBD genes in the same subclade have similar gene structures (Fig. 3B). The exon numbers of BrrLBD gene family members vary from one to three. Fourteen BrrLBD genes have only one exon (24%), most members of BrrLBD gene family have two exons (71%), and only three BrrLBD genes have three exons. The MEME online tool was performed to predict the conserved motif composition of turnip LBD proteins (Fig. 3C). The numbers of BrrLBD protein motifs range from 2 to 8. The motifs of BrrLBDs were annotated using InterProScan. The conserved CX2CX6CX3C zinc finger-like domain sequence was detected in motif 2, which exists in various species in all known LBD proteins. Furthermore, the leucine zipper-like coiled-coil was found in motif 1 and 3, which exists only in majority of the class Ⅰ LBD proteins (Majer and Hochholdinger, 2011; Shuai et al., 2002).
|
| Fig. 3 The gene structure and motif composition of BrrLBD genes (3A) Phylogenetic tree and classification of BrrLBD proteins was constructed by MEGA 7.0.21. (3B) Gene structure of BrrLBD genes. The yellow boxes and black lines represent the CDS and intron. (3C) Conserved motif compositions of BrrLBD proteins were identified using MEME. Each color represents a specific motif. |
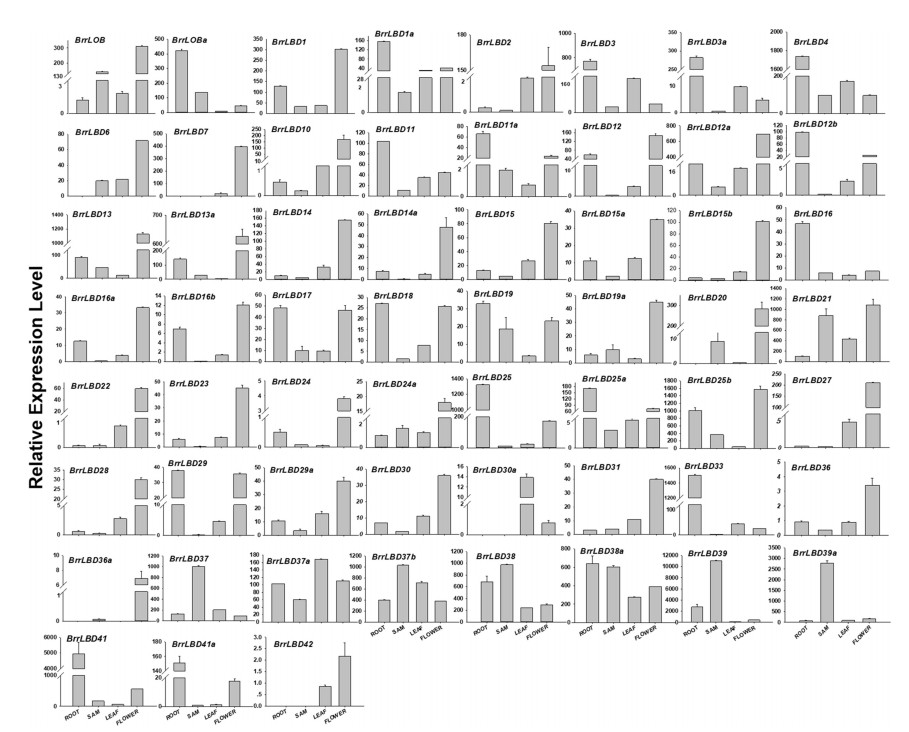
The gene expression pattern is always relative to its function (Xu et al., 2015). To investigate possible functions of BrrLBD genes in turnip, quantitative real-time fluorescence PCR (qRT-PCR) was performed to examine expression levels of 59 BrrLBD genes in different turnip tissues (Fig. 4). Except for BrrLBD30a, which is expressed specifically in leaves, 14 of 49 class Ⅰ BrrLBD genes are highly expressed in roots, 34 are strongly expressed in flower buds, and all class Ⅰ BrrLBD genes are weakly expressed in leaves.
|
| Fig. 4 Expression patterns of BrrLBD genes in root, shoot apical meristem (SAM), leaf, and flower buds (Flower) All data obtained from quantitative real-time fluorescence PCR. Constitutive β-tubulin gene served as an internal control. The error bar represents standard deviations for three technical duplications. |
Tissue-specific expression patterns are similar for genes within each phylogenetic subclade. For instance, all BrrLBD genes in class Ib and class Ⅰe, 12 of 16 BrrLBD genes in class Ⅰc, and 7 of 10 class Ⅰd genes are expressed highly in flower-buds. However, class Ⅰa genes showed more widespread but less tissue-specific expression patterns; for example, BrrLBD1, 12, 12a, 23, 24 and 24a are highly expressed in flower-buds, and BrrLBD1a, 3, 3a, 4, 11, 11a and 12b are expressed highly in roots. Seventeen duplicated BrrLBD pairs (BrrLBD11/11a, BrrLBD12/12a, BrrLBD13/13a, BrrLBD14/14a, BrrLBD15/15b, BrrLBD23/24, BrrLBD25/25a, BrrLBD38/38a, BrrLBD39/ 39a and BrrLBD41/41a) have similar tissue-specific expression patterns, respectively. In contrast, BrrLOB/BrrLOBa, BrrLBD16/16a, BrrLBD19/19a, BrrLBD29/29a, BrrLBD30/30a and BrrLBD37/37a have different tissue expression characteristics. These results imply that most turnip LBD gene functions have been conserved, although some gene functions may have been altered through gene duplication events.
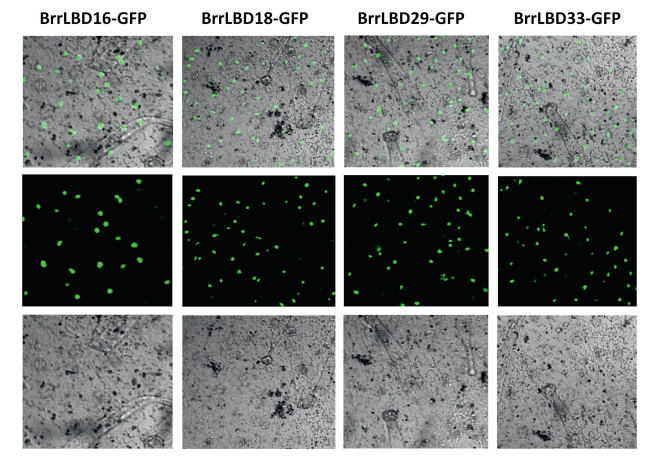
Subcellular localizationThe known members of the LBD gene family function as transcription factors that regulate plant-specific process. The GFP gene was fused with BrrLBDs as a reporter. The GFP signals of BrrLBD-GFP were detected in the nucleus of tobacco leaf epidermal cells (Fig. 5), which suggests that BrrLBD16, BrrLBD18, BrrLBD29, and BrrLBD33 might function as transcription factors.
|
| Fig. 5 Subcellular localization of 35S:BrrLBD-GFP in Nicotiana benthamiana leaves BrrLBD16-GFP, BrrLBD18-GFP, BrrLBD29-GFP and BrrLBD33-GFP were localized in the nucleus. |
LBD proteins belong to a novel family of plant-specific transcription factors involved in the regulation of processes during plant development and growth in higher plants, such as pollen development, plant regeneration, pathogen response, secondary growth, photomorphogenesis, pulvinus identity and petiole development (Xu et al., 2016). Turnip underwent polyploidization events, such as γ triplication (135 MYA) and β (90-100 MYA) and a (24-40 MYA) duplications. Following these polyploidization events, the Brassica genome underwent chromosome reduction and rearrangement and numerous gene losses took place, resulting in highly complex gene families (Du et al., 2017). Phylogenetic analysis of LBD proteins in apple, grape, eucalyptus, rice and Arabidopsis have led to the division of LBD families into two classes (class Ⅰ and class Ⅱ) (Cao et al., 2016; Lu et al., 2017; Wang et al., 2013; Yang et al., 2006). In this study, 59 BrrLBD genes were identified from turnip genome sequences, which were distributed on 10 chromosomes. Some BrrLBD proteins were detected in the nucleus. An unrooted phylogenetic tree divided BrrLBDs into 7 subclades (subclade a to e within class Ⅰ, and subclade a and b within class Ⅱ). Alignment analysis revealed that in turnip genome 7 AtLBD genes had no orthologs and 17 AtLBDs had more than one ortholog. These results demonstrate that the expanded of BrrLBD gene family in turnip may have been involved in chromosomal duplication events, containing multiple segmental duplication, tandem duplications, transposition events and genome-wide duplications.
Structural analysis is another effective method to mine the evolutionary history of the gene duplication events and phylogenetic relationship within a gene family. In this investigation, most BrrLBD genes had a similar exon/intron structure within the same subclade, which is consistent with LBD genes in A. thaliana, Oryza sativa, Eucalyptus grandis, Vitis vinifera and Malus domestica (Cao et al., 2016; Lu et al., 2017; Shuai et al., 2002; Wang et al., 2013; Yang et al., 2006). Motif analysis demonstrated that most BrrLBD proteins possessed similar motif distributions within the same subclade. The characteristic LOB domain was discovered in BrrLBD proteins, which consists of a CX2CX6CX3C zinc finger-like motif required for the DNA-binding activity within motif 2, a Gly-Ala-Ser (GAS) Block within motif 1, and a leucine-zipper-like coiled-coil motif responsible for protein interactions within motif 1 and 3. The zinc finger-like motif and GAS block were discovered in all 59 BrrLBDs. However, the leucine-zipper-like motif was discovered in 47 of 49 BrrLBDs within class Ⅰ and all remaining BrrLBD proteins contained an incomplete structure, which is similar to other species (Lu et al., 2017; Majer and Hochholdinger, 2011; Shuai et al., 2002; Xu et al., 2016). These results suggest that the gene structure and protein motif compositions of LBD genes are relatively conserved in different angiosperms.
In Arabidopsis, previous studies have demonstrated that LBD genes participate in plant growth and development as response factors and regulate nitrogen metabolism and anthocyanin synthesis as repressors (Kim and Lee, 2013; Mangeon et al., 2011; Okushima et al., 2007; Rubin et al., 2009). One important indicator of gene function is its expression profile. In this study, we detected the expression profiles of 59 BrrLBD genes in different tissues using qRT-PCR. These genes displayed distinct expression profiles in different turnip organs. Meanwhile, most genes exhibited similar expression profiles within each subclade. Most (10 of 17) duplicated BrrLBD protein pairs demonstrated similar tissuespecific expression patterns. In contrast, the remaining 7 gene duplicated pairs showed different tissue expression characteristics. These results imply that most gene functions have been conserved, although some gene functions may have changed during gene duplication events. Furthermore, the expression of BrrLBD33 in roots suggests that this gene may play a role in the development of the lateral root in turnip.
Declaration of Competing InterestThe authors declare no conflict of interests.
AcknowledgmentsThis study was supported by the Major Program of National Natural Science Foundation of China, China (31590820 and 31590823), the National Natural Science Foundation of China, China (41771123 and 31400244) and the Natural Science Foundation of Yunnan Province (2017FB050).
Appendix A. Supplementary dataSupplementary data to this article can be found online at https://doi.org/10.1016/j.pld.2019.11.004.
Artimo P., Jonnalagedda M., Arnold K., Baratin D., Csardi G., de Castro E., Duvaud S., Flegel V., Fortier A., Gasteiger E., et al, 2012. ExPASy:SIB bioinformatics resource portal. Nucleic Acids Res, 40: W597-W603. DOI:10.1093/nar/gks400 |
Bailey T.L., Elkan C., 1994. Fitting a mixture model by expectation maximization to discover motifs in biopolymers. Proc Int Conf Intell Syst Mol Biol, 2: 28-36. |
Cabrera J., Diaz-Manzano F.E., Sanchez M., Rosso M.N., Melillo T., Goh T., Fukaki H., Cabello S., Hofmann J., Fenoll C., et al, 2014. A role for LATERAL ORGAN BOUNDARIES-DOMAIN 16 during the interaction Arabidopsis-Meloidogyne spp. provides a molecular link between lateral root and root-knot nematode feeding site development. New Phytol, 203: 632-645. DOI:10.1111/nph.12826 |
Cao H., Liu C.Y., Liu C.X., Zhao Y.L., Xu R.R., 2016. Genomewide analysis of the lateral organ boundaries domain gene family in Vitis vinifera. J. Genet, 95: 515-526. DOI:10.1007/s12041-016-0660-z |
Chen J., Moreau C., Liu Y., Kawaguchi M., Hofer J., Ellis N., Chen R., 2012. Conserved genetic determinant of motor organ identity in Medicago truncatula and related legumes. Proc. Natl. Acad. Sci. USA, 109: 11723-11728. DOI:10.1073/pnas.1204566109 |
Cortizo M., Laufs P., 2012. Genetic basis of the "sleeping leaves" revealed. Proc. Natl. Acad. Sci. USA, 109: 11474-11475. DOI:10.1073/pnas.1209532109 |
Du J., Hu S., Yu Q., Wang C., Yang Y., Sun H., Yang Y., Sun X., 2017. Genome-wide identification and characterization of BrrTCP transcription factors in Brassica rapa var. rapa. Front. Plant Sci, 8: 1588. DOI:10.3389/fpls.2017.01588 |
Fan M., Xu C., Xu K., Hu Y., 2012. LATERAL ORGAN BOUNDARIES DOMAIN transcription factors direct callus formation in Arabidopsis regeneration. Cell Res, 22: 1169-1180. DOI:10.1038/cr.2012.63 |
Feng Z., Sun X., Wang G., Liu H., Zhu J., 2012. LBD29 regulates the cell cycle progression in response to auxin during lateral root formation in Arabidopsis thaliana. Ann. Bot, 110: 1-10. DOI:10.1093/aob/mcs019 |
Gaut B.S., Morton B.R., McCaig B.C., Clegg M.T., 1996. Substitution rate comparisons between grasses and palms:synonymous rate differences at the nuclear gene Adh parallel rate differences at the plastid gene rbcL. Proc. Natl. Acad. Sci. USA, 93: 10274-10279. DOI:10.1073/pnas.93.19.10274 |
Guo B.J., Wang J., Lin S., Tian Z., Zhou K., Luan H.Y., Lyu C., Zhang X.Z., Xu R.G., 2016. A genome-wide analysis of the ASYMMETRIC LEAVES2/LATERAL ORGAN BOUNDARIES (AS2/LOB) gene family in barley (Hordeum vulgare L.). J. Zhejiang Univ.-Sci. B, 17: 763-774. DOI:10.1631/jzus.B1500277 |
Hu B., Jin J., Guo A.Y., Zhang H., Luo J., Gao G., 2015. GSDS 2. 0:an upgraded gene feature visualization server. Bioinformatics, 31: 1296-1297. DOI:10.1093/bioinformatics/btu817 |
Hu Y., Zhang J., Jia H., Sosso D., Li T., Frommer W.B., Yang B., White F.F., Wang N., Jones J.B., 2014. Lateral organ boundaries 1 is a disease susceptibility gene for citrus bacterial canker disease. Proc. Natl. Acad. Sci. USA, 111: E521-E529. DOI:10.1073/pnas.1313271111 |
Iwakawa H., Ueno Y., Semiarti E., Onouchi H., Kojima S., Tsukaya H., Hasebe M., Soma T., Ikezaki M., Machida C., et al, 2002. The ASYMMETRIC LEAVES2 gene of Arabidopsis thaliana, required for formation of a symmetric flat leaf lamina, encodes a member of a novel family of proteins characterized by cysteine repeats and a leucine zipper. Plant Cell Physiol, 43: 467-478. DOI:10.1093/pcp/pcf077 |
Kim J., Lee H.W., 2013. Direct activation of EXPANSIN14 by LBD18 in the gene regulatory network of lateral root formation in Arabidopsis. Plant Signal. Behav, 8: e22979. DOI:10.4161/psb.22979 |
Kim M.J., Kim M., Lee M.R., Park S.K., Kim J., 2015. LATERAL ORGAN BOUNDARIES DOMAIN (LBD)10 interacts with SIDECAR POLLEN/LBD27 to control pollen development in Arabidopsis. Plant J.:Cell Mol Biol, 81: 794-809. DOI:10.1111/tpj.12767 |
Kumar S., Stecher G., Tamura K., 2016. MEGA7:molecular evolutionary genetics analysis version 7. 0 for bigger datasets. Mol. Biol. Evol, 33: 1870-1874. DOI:10.1093/molbev/msw054 |
Lee H.W., Kang N.Y., Pandey S.K., Cho C., Lee S.H., Kim J., 2017. Dimerization in LBD16 and LBD18 transcription factors is critical for lateral root formation. Plant Physiol, 174: 301-311. DOI:10.1104/pp.17.00013 |
Lee H.W., Kim M.J., Park M.Y., Han K.H., Kim J., 2013. The conserved proline residue in the LOB domain of LBD18 is critical for DNA-binding and biological function. Mol. Plant, 6: 1722-1725. DOI:10.1093/mp/sst037 |
Lin K., Zhang N., Severing E.I., Nijveen H., Cheng F., Visser R.G., Wang X., de Ridder D., Bonnema G., 2014. Beyond genomic variation-comparison and functional annotation of three Brassica rapa genomes:a turnip, a rapid cycling and a Chinese cabbage. BMC Genomics, 15: 250. DOI:10.1186/1471-2164-15-250 |
Liu H., Wang S., Yu X., Yu J., He X., Zhang S., Shou H., Wu P., 2005. ARL1, a LOB-domain protein required for adventitious root formation in rice. Plant J, 43: 47-56. DOI:10.1111/j.1365-313X.2005.02434.x |
Liu J., Sheng L., Xu Y., Li J., Yang Z., Huang H., Xu L., 2014. WOX11 and 12 are involved in the first-step cell fate transition during de novo root organogenesis in Arabidopsis. Plant Cell, 26: 1081-1093. DOI:10.1105/tpc.114.122887 |
Lu Q., Shao F., Macmillan C., Wilson I.W., van der Merwe K., Hussey S.G., Myburg A.A., Dong X., Qiu D., 2017. Genomewide analysis of the lateral organ boundaries domain gene family in Eucalyptus grandis reveals members that differentially impact secondary growth. Plant Biotechnol. J, 16: 124-136. |
Luo Y., Ma B., ZengQ., XiangZ., He N., 2016. Identification and characterizationof lateral organ boundaries domain genes in mulberry, Morus notabilis. Meta gene, 8: 44-50. DOI:10.1016/j.mgene.2014.04.004 |
Majer C., Hochholdinger F., 2011. Defining the boundaries:structure and function of LOB domain proteins. Trends Plant Sci, 16: 47-52. |
Majer C., Xu C., Berendzen K.W., Hochholdinger F., 2012. Molecular interactions of ROOTLESS CONCERNING CROWN AND SEMINAL ROOTS, a LOB domain protein regulating shoot-borne root initiation in maize (Zea mays L.). Philos. Trans. R. Soc. Lond. Ser. B Biol. Sci, 367: 1542-1551. DOI:10.1098/rstb.2011.0238 |
Mangeon A., Bell E.M., Lin W.C., Jablonska B., Springer P.S., 2011. Misregulation of the LOB domain gene DDA1 suggests possible functions in auxin signalling and photomorphogenesis. J. Exp. Bot, 62: 221-233. DOI:10.1093/jxb/erq259 |
Oh S.A., Park K.S., Twell D., Park S.K., 2010. The SIDECAR POLLEN gene encodes a microspore-specific LOB/AS2 domain protein required for the correct timing and orientation of asymmetric cell division. Plant J.:Cell Mol Biol, 64: 839-850. DOI:10.1111/j.1365-313X.2010.04374.x |
Okushima Y., Fukaki H., Onoda M., Theologis A., Tasaka M., 2007. ARF7 and ARF19 regulate lateral root formation via direct activation of LBD/ASL genes in Arabidopsis. Plant Cell, 19: 118-130. DOI:10.1105/tpc.106.047761 |
Rubin G., Tohge T., Matsuda F., Saito K., Scheible W.R., 2009. Members of the LBD family of transcription factors repress anthocyanin synthesis and affect additional nitrogen responses in Arabidopsis. Plant Cell, 21: 3567-3584. DOI:10.1105/tpc.109.067041 |
Shuai B., Reynaga-Pena C.G., Springer P.S., 2002. The lateral organ boundaries gene defines a novel, plant-specific gene family. Plant Physiol, 129: 747-761. DOI:10.1104/pp.010926 |
Thatcher L.F., Powell J.J., Aitken E.A., Kazan K., Manners J.M., 2012. The lateral organ boundaries domain transcription factor LBD20 functions in Fusarium wilt Susceptibility and jasmonate signaling in Arabidopsis. Plant Physiol, 160: 407-418. DOI:10.1104/pp.112.199067 |
Wang X., Zhang S., Su L., Liu X., Hao Y., 2013. A genome-wide analysis of the LBD (LATERAL ORGAN BOUNDARIES domain) gene family in Malus domestica with a functional characterization of MdLBD11. PLoS One, 8: e57044. DOI:10.1371/journal.pone.0057044 |
Xu C., Luo F., Hochholdinger F., 2016. LOB domain proteins:beyond lateral organ boundaries. Trends Plant Sci, 21: 159-167. |
Xu Z., Sun L., Zhou Y., Yang W., Cheng T., Wang J., Zhang Q., 2015. Identification and expression analysis of the SQUAMOSA promoter-binding protein (SBP)-box gene family in Prunus mume. Mol. Genet. Genom.:MGG, 290: 1701-1715. DOI:10.1007/s00438-015-1029-3 |
Yang Y., Yu X., Wu P., 2006. Comparison and evolution analysis of two rice subspecies LATERAL ORGAN BOUNDARIES domain gene family and their evolutionary characterization from Arabidopsis. Mol. Phylogenetics Evol, 39: 248-262. DOI:10.1016/j.ympev.2005.09.016 |
Yang Z., 2007. PAML 4:phylogenetic analysis by maximum likelihood. Mol. Biol. Evol, 24: 1586-1591. DOI:10.1093/molbev/msm088 |
Yordanov Y.S., Regan S., Busov V., 2010. Members of the LATERAL ORGAN BOUNDARIES DOMAIN transcription factor family are involved in the regulation of secondary growth in Populus. Plant Cell, 22: 3662-3677. DOI:10.1105/tpc.110.078634 |
Zhang Y.M., Zhang S.Z., Zheng C.C., 2014. Genomewide analysis of LATERAL ORGAN BOUNDARIES Domain gene family in Zea mays. J. Genet, 93: 79-91. |
Zhou C., Han L., Fu C., Chai M., ZhangW., Li G., Tang Y., Wang Z.Y., 2012. Identification and characterization of petiolule-like pulvinus mutants with abolished nyctinastic leafmovement in themodel legume Medicago truncatula. New Phytol, 196: 92-100. DOI:10.1111/j.1469-8137.2012.04268.x |



