b. Institute of Plant Breeding, Genetics and Genomics(Department of Crop and Soil Sciences), and Department of Plant Biology, University of Georgia, Athens, GA 30602, USA;
c. Laboratory of Bioaggressors and Integrated Protection in Agriculture, The National Agronomic Institute of Tunisia, University of Carthage, 43 Avenue Charles-Nicolle, Tunis 1082, Tunisia;
d. Anand Agricultural University, Anand, India;
e. USDA-ARS National Clonal Germplasm Repository for Citrus and Dates, Riverside, CA, USA
Date palm (Phoenix dactylifera) is a keystone tree species in the oasis agrosystems of North Africa and Southwestern Asia (Tengberg, 2012; Terral et al., 2012). Date palms are a key part of the history and culture of these regions. The genus Phoenix L. includes 14 species (Govaerts and Dransfield, 2005) traditionally distributed in the Old World from the Canary and Cape Verde islands in the Atlantic Ocean, throughout Africa, Madagascar and Asia, reaching Sumatra, Taiwan (China) and the Philippines in the East. There are very few studies on the phylogenetic analysis of Phoenix and each of them provided different answers (Barrow, 1998; Pintaud et al., 2010; Pintaud et al., 2013; Torres et al., 2018). A consensus based on the chloroplastic loci support a P. dactylifera clade, which includes P. dactylifera, Phoenix sylvestris, and Phoenix atlantica (Pintaud et al., 2010). However, phylogenetic analysis based on individual nuclear genes (e.g. CYP703, LOG, cytidine deaminase) from 14 Phoenix species suggested a P. dactylifera subclade with P. dactylifera, Phoenix theophresti, and P. atlantica (Torres et al., 2018). So far, there is no comprehensive phylogenetic analysis based on either whole organellar genome assembly or whole nucleargenome assemblies of all members of the Phoenix genus.
The primary center of diversity for Phoenix is an area from India to Indochina, where eight species are found (Pintaud et al., 2010). According to archaeological data, date palm was first cultivated in ~5000-3000 BC near the Persian Gulf, and quickly spreading to the countries that are now called Iran, Iraq and the United Arab Emirates (UAE) (Tengberg, 2012; Abbo et al., 2015). The spread of date palms is believed to have occurred over many centuries along two main routes, from Iraq east to Iran, Pakistan and India and west from Egypt to North Africa (Erskine et al., 2004). However, recent analyses of date palm diversity have suggested that North Africa may have been the origin point of domesticated date palm, with subsequent dissemination east to Asia (Hazzouri et al., 2015). European exploration and colonization have spread date palm throughout much of the world. There is a growing interest in the genetics and genomics of date palm because of its exceptional contributions to the livelihoods of desert farming communities in Asia and North Africa, and also because of its potential as a source for valuable agronomic and stress-tolerance traits. Most of the wild date palm germplasm is already lost, and only a few natural populations are believed to exist (Gros-Balthazard et al., 2017; Wales and Blackman, 2017). Genetic diversity existing in the cultivated date palm is also being lost because of shifts to fewer modern varieties (El-Juhany, 2010). This will increase the vulnerability of date palm to sudden changes in climate, diseases (e.g., Bayoud, and lethal yellowing) and insect pests (e.g., red palm weevil).
Date palms have a juvenile phase of 5-8 years and life spans of over 50 years. Superior cultivars are propagated and distributed by vegetative propagation from offshoots or tissue culture. Hence, though date palm domestication is quite ancient, it is a relatively recent event on a generation time scale, in comparison to annual crops. Modern date palm improvement would be facilitated greatly by a genetic description of the diversity that is present worldwide and the correlation of particular desired traits with specific markers (Morrell et al., 2012). The value of genetic diversity to modern plant breeding is enormous. Such important traits as improved yield, disease and abiotic stress resistance, improved fruit quality, and longer shelf life have been successfully transferred from landraces and wild germplasm to elite cultivated varieties in several crop species. In spite of the many potential benefits of wild Phoenix germplasm, there is no reported effort on the phenotypic evaluation of the wild relatives and hybridization of cultivated date palm with other Phoenix species. Also, an important date palm-specific problem is the difficulty in identifying cultivars until the fruit is produced. Thus, efficient assessment of genetic composition in date palm requires markers that differentiate similar-looking cultivars. Moreover, these markers should be able to identify the genotypes of new varieties with desirable agronomic traits that may have emerged spontaneously in remote (e.g., oasis) locations through sexual reproduction. Studies of genetic diversity and gene flow in date palm can help to devise targeted approaches to conserve germplasm diversity and to investigate evolutionary processes within the genus.
Microsatellites, otherwise known as Simple Sequence Repeats (SSRs), have been used to assess genetic diversity and relatedness of date palm varieties in Algeria (Akkak et al., 2009), Iran (Arabnezhad et al., 2012), Iraq (Jubrael et al., 2005), Tunisia (Hamza et al., 2011, 2012; Zehdi-Azouzi et al., 2015), Saudi Arabia (Al-Abdoulhadi et al., 2011), Sudan (Elshibli and Korpelainen, 2008) Oman (Al-Ruqaishi et al., 2008), Qatar (Ahmed and Al-Qaradawi, 2009; Elmeer and Mattat, 2015), UAE (Chaluvadi et al., 2014), Libya (Racchi et al., 2014), and Morocco (Sedra, 2010). Recent studies have looked into the genetic diversity of worldwide date palm germplasms, using either SSRs (Zehdi-Azouzi et al., 2015), single nucleotide polymorphisms (SNPs) (Hazzouri et al., 2015; Mathew et al., 2015) or comparisons of whole genomes (Hazzouri et al., 2015). Each of these studies investigated different sets of accessions (with some common genotypes) and they used different tools to assess diversity. In general, these broad germplasm investigations identified separate germplasms for Asian and Africa accessions, and some structure within these major subgroups. Further analyses are needed to investigate the complete germplasm collection for this important desert crop and its wild relatives.
The principal aim of this work was to resolve the phylogeny of the genus Phoenix using whole plastome assemblies of all Phoenix species and to analyze a broad distribution of cultivated date palm and related species collected from 13 countries to evaluate the genetic diversity and structure of date palm germplasm. These results were analyzed to understand better the relationship between date palm accessions collected from North Africa and Asia, and to assess the roles of biogeographical history and human activity on the current diversity and distribution of date palm germplasm.
2. Methods 2.1. Chloroplast genome assembly, annotation and phylogenetic analysisGenomic shotgun sequence data were obtained from 27 Phoenix accessions representing 14 Phoenix species, available in NCBI Genbank as a part of a previous study (Torres et al., 2018) (Supplementary Table 1). The reference plastomes assemblies of P. dactylifera cv. Khalas (GenBank: NC_013991.2) and cv. Aseel (GenBank: FJ212316) were also included in this analysis. This study included three important accessions of P. dactylifera, Khalas, Deglet Noor and Aseel, which are major cultivars in Saudi Arabia, North Africa and Pakistan, respectively. Plastid-homologous sequences were selected from raw Illumina sequence data of Phoenix accessions and then de novo assembled using Velvet and further scaffolding in Geneious 10.1.2, as previously described (Frailey et al., 2018; Vaughn et al., 2014). Plastome assemblies were annotated using the program DOGMA (Wyman et al., 2004). Protein-coding regions, rRNAs, tRNAs, introns, and intergenic regions were all annotated and extracted from DOGMA. A total of 52 gene sequences shared by all Phoenix accessions and one Elaeis guineensis (oil palm) accession were individually aligned using ClustalW (Larkin et al., 2007) in Geneious 10.1.2 (https://www.geneious.com). The individual genes used in the phylogenetic analyses were accD, atpB, atpF, ccsA, infA, ndhA, ndhC, ndhD, ndhF, ndhG, ndhH, ndhI, ndhK, orf188, petA, petB, petD, petG, petL, petN, psaA, psaB, psaC, psaI, psaJ, psbE, psbG, psbH, psbI, psbJ, psbK, psbL, psbN, psbZ, rbcL, rpl14, rpl16, rpl20, rpl22, rpl32, rpl33, rpl36, rpoA, rpoC1, rpoC2, rps2, rps3, rps4, rps8, rps11, rps15, and rps16. Identically aligned regions of each gene sequence were extracted, concatenated, and realigned using ClustalW in Geneious 10.1.2. The E. guineensis plastome was utilized as the outgroup. The Bayesian phylogenetic analysis was done on MrBayes 3.2.1 (Ronquist et al., 2012), as reported in a previous study (Frailey et al., 2018). A separate phylogenetic analysis was also carried out using the alignment of whole plastome assemblies of all the Phoenix species. The whole plastome assemblies were aligned using progressiveMauve aligner, which accurately aligns colinear sequences even if they have undergone large numbers of nucleotide substitutions, indels and rearrangements (Darling et al., 2004, 2010).
| Locus | Alleles | He | Ho | FST | |
| 1 | DP151 | 48 | 0.87 | 0.15 | 0.48 |
| 2 | DP152 | 44 | 0.72 | 0.34 | 0.53 |
| 3 | DP153 | 39 | 0.66 | 0.30 | 0.53 |
| 4 | DP154 | 49 | 0.84 | 0.46 | 0.45 |
| 5 | DP159 | 50 | 0.89 | 0.56 | 0.37 |
| 6 | DP160 | 52 | 0.85 | 0.58 | 0.32 |
| 7 | DP164 | 47 | 0.86 | 0.56 | 0.34 |
| 8 | DP167 | 35 | 0.82 | 0.45 | 0.45 |
| 9 | DP165 | 34 | 0.71 | 0.45 | 0.37 |
| 10 | DP170 | 43 | 0.85 | 0.75 | 0.12 |
| 11 | DP171 | 35 | 0.75 | 0.39 | 0.48 |
| 12 | DP172 | 23 | 0.64 | 0.67 | -0.06 |
| 13 | JLBDP9 | 27 | 0.76 | 0.56 | 0.78 |
| 14 | JLBDP15 | 58 | 0.91 | 0.17 | 0.38 |
| 15 | JLBDP12 | 61 | 0.94 | 0.51 | 0.45 |
| 16 | JLBDP20 | 57 | 0.94 | 0.66 | 0.29 |
| 17 | DPG1229 | 36 | 0.78 | 0.28 | 0.64 |
| 18 | DPG2058 | 32 | 0.74 | 0.33 | 0.55 |
| 19 | DPG2216 | 30 | 0.76 | 0.85 | 0.43 |
| Mean | 42.1 | 0.80 | 0.47 | 0.42 | |
| *He = Expected heterozygosity, Ho = Observed heterozygosity, FST = Fixation index. | |||||
The initial date palm collection for our population genetic analyses consisted of DNA samples from 210 cultivated accessions of P. dactylifera and 16 accessions of other Phoenix species from 13 countries. These samples include 112 accessions collected and maintained by the United States Department of Agriculture - National Clonal Germplasm Repository for Citrus & Dates (USDA-NCGRCD) in California, 81 samples collected and maintained at research stations in UAE, 12 accessions collected from Tunisia, 19 accessions collected from India and two accessions collected from Pakistan (Supplementary Table 2). DNAs were isolated from leaf samples using DNeasy plant mini kits as per manufacturer's (Qiagen, USA) instructions. The DNA sample representing each accession was extracted from a single leaf of a single plant of that cultivar. Multiple (commonly three) trees were independently sampled for each accession grown in the California USDA collection (Supplementary Table 2).
| Country | Number of samples | N | Na | Ne | I | He | Ho | uHe |
| Morocco | 11 | 10.79 | 7.11 | 4.43 | 1.55 | 0.7 | 0.51 | 0.74 |
| Algeria | 16 | 15.63 | 10.74 | 6.94 | 1.97 | 0.8 | 0.55 | 0.83 |
| Tunisia | 13 | 12.42 | 8.21 | 5.44 | 1.8 | 0.78 | 0.32 | 0.82 |
| Egypt | 11 | 9.84 | 8.21 | 5.76 | 1.8 | 0.78 | 0.52 | 0.82 |
| Sudan | 4 | 4 | 3.47 | 3.06 | 1.04 | 0.57 | 0.47 | 0.65 |
| Iraq | 42 | 41.21 | 18.68 | 7.12 | 2.2 | 0.81 | 0.6 | 0.82 |
| Saudi Arabia | 16 | 15.53 | 10 | 5.91 | 1.9 | 0.79 | 0.47 | 0.81 |
| UAE | 21 | 10.74 | 8 | 4.98 | 1.74 | 0.75 | 0.44 | 0.79 |
| Oman | 15 | 14.89 | 8.79 | 5.64 | 1.84 | 0.79 | 0.46 | 0.82 |
| Pakistan | 2 | 2 | 2.21 | 1.99 | 0.65 | 0.41 | 0.34 | 0.54 |
| India | 16 | 15.89 | 9.42 | 4.68 | 1.69 | 0.69 | 0.37 | 0.72 |
| USA | 16 | 16 | 10.89 | 6.42 | 1.99 | 0.81 | 0.57 | 0.83 |
| *N = No. of alleles, Na = No. of different alleles, Ne = No. of effective alleles, I = Shannon's information index, Ho = Observed heterozygosity, He = Expected heterozygosity, uHe = Unbiased expected heterozygosity, FST = Fixation index. | ||||||||
Nineteen polymorphic nuclear SSR loci (Elmeer et al., 2011, Zhao et al., 2012) (Supplementary Table 3) were amplified with polymerase chain reaction (PCR) performed using a three-primer system with an M13 universal fluorescent-labeled primer (FAM, HEX, NED), as described previously (Chaluvadi et al., 2014). The PCR products were detected and sized on an ABI 3730 sequencer (Applied Biosystems, Foster City, CA). Resultant chromatograms were scored using Soft Genetics Genemarker Version 2.4.
2.4. Population genetic analysesOf the 226 accessions investigated, 195 accessions (which included 182 cultivated varieties/landraces of P. dactylifera and 13 accessions of other Phoenix species) were chosen for full analysis. The accessions were chosen because they yielded less than 11% null alleles with 19 microsatellite loci, and they also reduced overrepresentation of accessions originating from Iraq and UAE in our initial sampling. Population genetic statistics were calculated using Identity (Wagner and Sefc, 1999), Microsat 2.0 (Minch et al., 1995) and GenAlEx (Peakall and Smouse, 2012). These analyses included characterization of allelic diversity, taxon-specific alleles, Shannon diversity index, expected heterozygosity (gene diversity), observed heterozygosity, and Wright's FST (Wright, 1950). We also tested deviations from HardyeWeinberg Equilibrium with GENEPOP version 4.5 (Rousset, 2008).
To partition the genetic variance based on the continent and country of origin, we analyzed molecular variance (AMOVA) (Excoffier et al., 1992). The fixation index FST was estimated with the overall dataset within each country, and among all pairs of populations. The statistical significance of FST estimates was tested by 1000 random permutations of individuals across populations using GeneAlEx. Nei's genetic distance (Nei and Chakraborty, 1973) and Chord genetic distance (Cavalli-Sforza and Edwards, 1967) were calculated between pairs of genotypes for use in cluster analysis and Principal Coordinates Analysis (PCoA).
The genetic structure of the population was studied using the model-based (Bayesian) clustering method implemented in software package STRUCTURE Ver 2.4.1 (Pritchard et al., 2000). Predefined numbers of populations (k) ranged from 2 to 15, with an initial burn-in period of 50, 000 replicates and 50, 000 Markov Chain Monte Carlo (MCMC) iterations. An accession was assigned to a cluster if the admixture coefficient was >80% (Qi > 0.8) for that group. Accessions with membership probabilities less than 0.8 were assigned to an admixture group (Diez et al., 2015; Stich et al., 2005). Ten independent simulations were run for each K value. We did not use prior information to define the clusters. Because these analyses require codominant alleles and are sensitive to missing data, only 19 microsatellites (each with fewer than 11% missing data) were used. The average K value was calculated from the ten runs and Delta K was calculated by a web-based program, Structure Harvester (Earl, 2012), to identify the number of populations that best reflect the population structure of our samples (Evanno et al., 2005). The STRUCTURE patterns chosen for display (Fig. 5 and Figure S1) were those with the highest statistical support from the ten independent runs. The multiple replicate runs from STRUCTURE were integrated with CLUMPP software (Jakobsson and Rosenberg, 2007).

|
| Fig. 1 Bayesian phylogenetic trees for (A) 52 shared chloroplast gene alignments and (B) whole plastome alignments from 29 Phoenix accessions representing 14 Phoenix species. Bayesian posterior probabilities are shown at the nodes. The scale indicates substitutions per site. The vertical lines labeled 1, 2 and 3 indicate three identified phylogenetic clusters. |
|
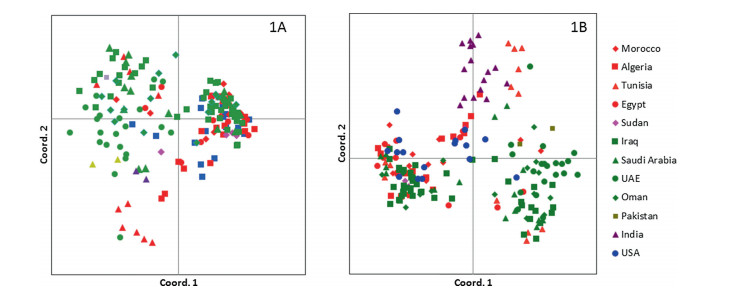
| Fig. 2 Scatterplot of Principal Coordinates Analysis (PCoA) produced using pairwise genetic distance matrix calculated in GenAlEx 6.5. The first two axes explain 20.4 and 15.4% of the total variation, respectively. Accessions are color-coded based on the original site of collection. Fig. 2A has two accessions from India, whereas Fig. 2B has 16 accessions from India. The samples listed as the USA are from the USDA-NCGRCD collection and do not have any information regarding where they were obtained (date palms are not native to the USA). |
|
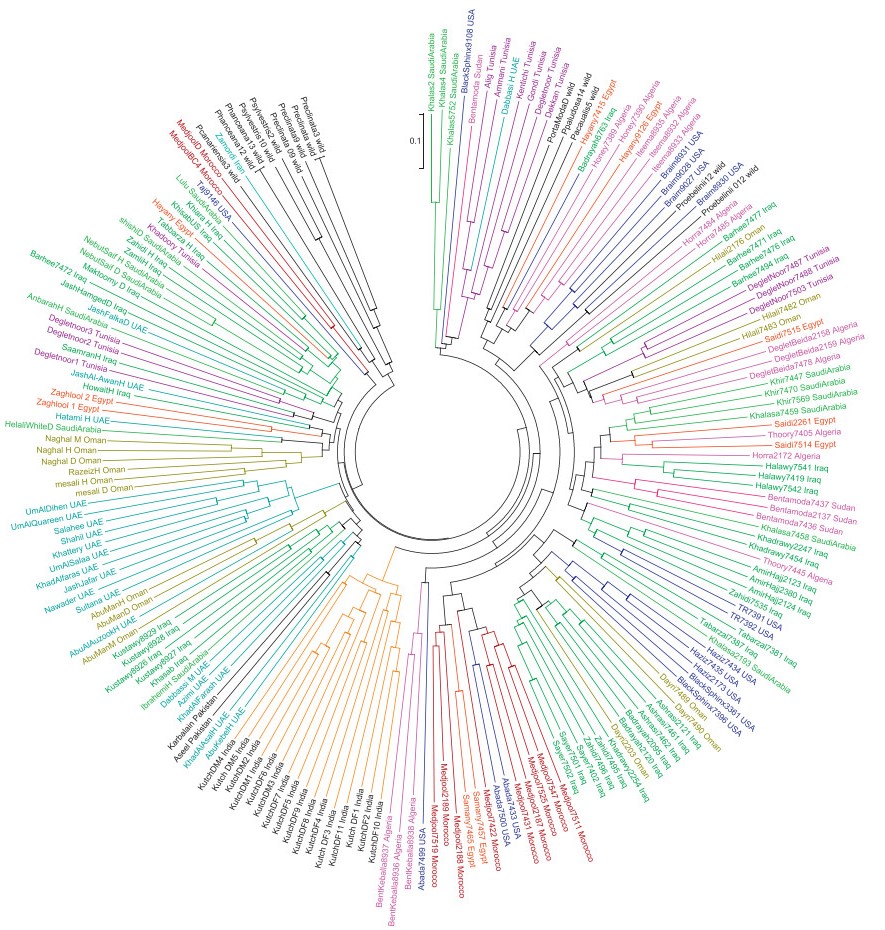
| Fig. 3 Minimum evolution tree based on chord genetic distances (Slatkin, 1995). The branches of the tree and accession names are color-coded based on the country of origin. |
|
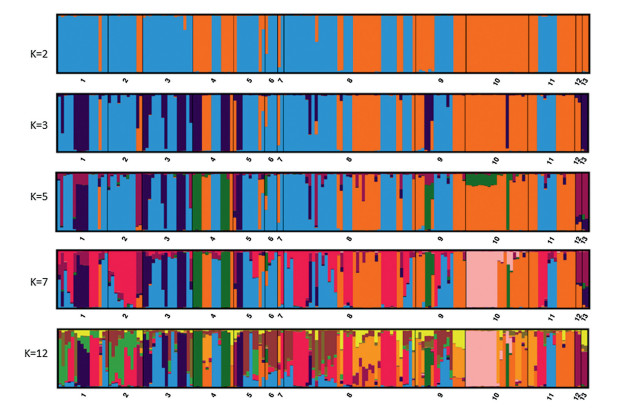
| Fig. 4 Model-based ancestry for each accession represented by a vertical bar partitioned into colored segments that represent the accession's estimated membership fractions. The accessions in the barplots for k = 2, k = 3, k = 5, k = 7 and k = 12 were arranged by countries of their origin, which include USA (1), Morocco (2), Algeria (3), Tunisia (4), Egypt (5), Sudan (6), Iran (7), Iraq (8), Saudi Arabia (9), UAE (10), Oman (11), Pakistan (12), India (13). |
|
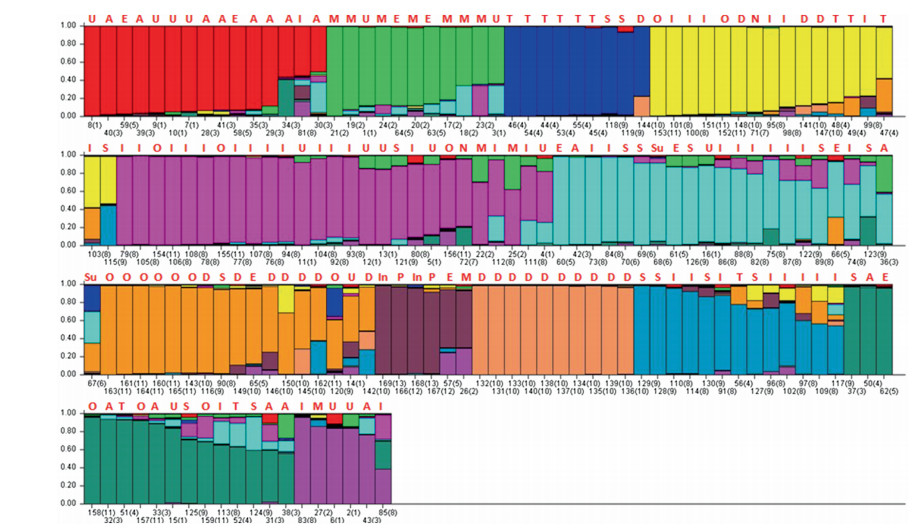
| Fig. 5 Structure barplot for k = 12. Colored segments denote the place of initial collection. Algeria (A), Morocco (M), Egypt (E), Sudan (Su), Tunisia (T), Iraq (I), Iran (N), India (In), Oman (O), Saudi Arabia (S), Pakistan (P) and UAE (D). Many of these samples were collected from the USDA-NCGRCD in the USA, but are labeled according to their original point of collection. The samples marked USA (U) are those from the USDA-NCGRCD that did not have information regarding their original site of collection. |
Full chloroplast genomes were assembled for most Phoenix species. Wherever we could not assemble, the gaps were filled with Ns. The missing regions in the plastomes of P. atlantica_female, Phoenix caespitosa_female, Phoenix canariensis_female, Phoenix paludosa_female, Phoenix pusilla_male, and Phoenix reclinata_female are likely less than 100 bp in total based on comparison to other sequences in this study. The plastome sizes of Phoenix species ranged from 156, 496 bp in P. reclinata_male to 160, 758 bp in Phoenix pauludosa_female. All the Phoenix genomes have 68-70 single copy genes, 18-20 duplicated genes, 31 tRNA genes and four rRNA genes. The whole plastome sequences of all the accessions are included in Supplementary Data. Sizes of all sequenced plastomes and their composition can be found in Supplementary Table 1. There were no major rearrangements in the sequenced plastomes relative to the P. dactylifera assembly.
Phylogenetic trees calculated by the maximum likelihood and Bayesian analyses were congruent in the overall topology, hence only trees calculated by Bayesian analysis using MrBayes are shown (Fig. 1A and B). The phylogenetic tree based on the alignments of only genes (Fig. 1A) has posterior probabilities (PP) of 0.95-1.0 for all nodes except the P. atlantica node. The plastome sequences of male and female accessions of each species were not identical. However, the male and female accessions of each species clustered as pairs in all the Phoenix species, except a few cases (Phoenix acaulis, Phoenix rupicola and Phoenix theophrasti), perhaps indicating a degree of sequence change since their divergence or mislabeling of some cultivars. Fig. 1A shows that the cultivated date palm accessions shared the same clade with male and female accessions of P. sylvestris, Phoenix caespitosa, P. atlantica and male accessions of P. acaulis and P. pusilla. The posterior probability of the P. atlantica branch was only 0.55 while all the other nodes in the phylogenetic tree were from 0.95 to 1.0. The date palm cultivar Aseel appeared at the base of the P. dactylifera clade. Fig. 1B provides the phylogenetic tree based on the whole plastome alignments. The branching structure of this phylogenetic tree is mostly in agreement with the chloroplast gene-based phylogenetic tree (Fig. 1A) with a few differences. The posterior probabilities of all the nodes are 1.0, except the node where P. reclinata (PP = 0.93) and the node where P. theophresti (PP = 0.93) diverged. All the male and female accessions of each species, except with P. acaulis, clustered together.
Based on the phylogenetic analyses of plastome sequences, we separated the phylogenetic trees into three groups. We call Group A the dactylifera group, which includes P. dactylifera, P. sylvestris, Phoenix acualis, P. caespitosa, P. pusilla and P. atlantica. Group B has P. rupicola, P. theophresti, P. canariensis, P. pauludosa, Phoenix roenbereni and P. reclinata. The third group, the most distantly related to P. dactylifera, consists of Phoenix hanceana and Phoenix andamanensis.
3.2. Genetic diversity analysisAn average of 42.1 alleles per locus was detected within the sample data (Table 1). The genetic variation in each accession and each locus, as estimated by the number of alleles, observed heterozygosity (Ho), expected heterozygosity (He) and fixation index (FST), are presented in Tables 1 and 2 and Supplementary Table 2. The values of expected heterozygosity ranged from 0.64 for the locus DP172 to 0.94 for the locus JLBDP20. The values of observed heterozygosity were lower than the value of expected heterozygosity at most of the loci (0.14-0.85). Values for the mean allelic diversity (richness) per locus ranged from 1.1 in date palm genotypes AbuMan, Dabbasi, Medjool and Shahil to 1.8 in date palm genotypes Ashrasi2121, BlackSphinx3361, Dayri7490, Halawy7419, Haziz7434, Haziz7435, Sayer7402, Zahidi749, Ashrasi7461, Ashrasi7462, Khadrawy2254, Khadrawy7454 and Sayer7502.
Unique alleles specific to each accession were identified for most of the accessions (Supplementary Table 2). Values for the mean heterozygosity ranged from 0.05 in date palm genotypes AbuMan, Dabbasi, Medjool and Shahil to 0.42 in date palm genotypes Ashrasi7461, Ashrasi7462, Khadrawy2254, Khadrawy7454 and Sayer7502 (Supplementary Table 1). The accessions of Iraqi origin have the highest allelic diversity and the highest number of private alleles (Table 2), but this may be largely because we had more accessions from Iraq (42) than from any other country (ranging from 2 to 20). We did not find any accessions that were 100% identical at all tested loci, even among an independent sampling of accessions with the same cultivar name.
Principal Coordinates Analysis was performed to visualize the relationships between the date palm accessions (Fig. 2A). The first two axes explained 20.4% and 15.4% of the variability, respectively. These two components separate the studied date palm accessions into two loose clusters with the remaining accessions scattered in between. One group contains accessions that were collected predominantly from Asia (UAE, Oman, Iraq, Saudi Arabia, Pakistan, and India), whereas the second group included accessions largely from North Africa and accessions of unknown origin from the USA. The data points scattered in between were mostly composed of accessions from Tunisia, thereby suggesting a particularly diverse and intermixed germplasm pool in this nation. Non-parametric AMOVA to find significant differences between continents produced a pvalue of 0.001. A separate analysis was conducted to include 16 accessions from India. The results were mostly in agreement with Fig. 2A except that the accessions collected from India formed a third cluster that was mostly related to the African germplasm (Fig. 2B).
From the accession genotyping data, we calculated pairwise chord genetic distances between all genotypes to generate a dissimilarity matrix from which a Weighted Neighbor-Joining tree was calculated. P. reclinata was used to root the phylogenetic tree. The relatedness tree based on chord genetic distances depicted in Fig. 3 showed 12 well-resolved clusters. The wild Phoenix species formed two clusters. One cluster consists of P. reclinata, P. sylvestris, P. hanceana and P. canariensis and one P. dactylifera accession from Iran. The other cluster contains Phoenix roebelenii, P. paludosa, Phoenix acaualis and some accessions from Algeria, Egypt, USA, and Iraq. Indian accessions formed a distinct cluster. The neighborjoining tree indicated that date palm accessions are not clustered primarily in a manner related to their country of origin. There are, however, country-specific clusters of accessions originating from India, Iraq, UAE, Tunisia and Morocco.
Most often, accessions with the same name, although collected from different locations, clustered together. However, some accessions with the same name but collected from different locations did not cluster together. For example, multiple accessions of Medjool and Deglet Noor were found in 2 and four different clusters, respectively. Moreover, we were able to identify alleles specific to each cultivar collected from a given location.
We observed few loci/country combinations that were in HWE (Supplementary Table 4), as expected given the history of natural and human selection acting on this crop. The mean FST value in the AMOVA analysis based on the average across continents and countries are 0.15 and 0.44 respectively. The hierarchical AMOVA revealed that the majority of total genetic variance (92%) was due to variation among accessions, while 8% was due to variation among countries. An analysis of genetic variation between Asia and Africa showed that 98% of the total genetic variance was due to the variation among accessions within each continent and only 2% of total variance was due to variation among continents (Supplementary Figure 1). We tested the significance of variance components by pairwise population comparisons of FST values using a non-parametric permutation approach (Excoffier et al., 1992). Supplementary Table 5 shows a matrix of the results for all the populations. Fifteen of 21 pairwise country comparisons found that the accessions originating from different countries were significantly different (p < 0.001).
3.3. Population structure analysisBayesian structure analysis followed by the delta K measure was used to estimate the number of sub-populations (Evanno et al., 2005). We observed the highest delta k peak at k = 2, which is followed by smaller peaks at k = 3, k = 5, k = 7, and k = 12. Fig. 4, derived from ten replicate runs from STRUCTURE integrated by CLUMPP software (Jakobsson and Rosenberg, 2007), shows the calculated accession ancestry at different k values. The samples are depicted by the approximate geography of origin, with African accessions on the left and Asian accessions on the right. At k = 2, each accession is color-coded with either African alleles (blue) or Asian alleles (orange). Fourteen of the 54 accessions originating from Africa have Asian alleles. The majority of these accessions with Asian alleles came from Tunisia. Similarly, 28 Asian accessions out of 99 have African alleles. The majority of these Asian accessions are from the collections originating from Iraq. At k = 3, we observed a distinct group with 17 accessions from Africa and five accessions from Asia.
At k = 12, when the STRUCTURE results are organized by degree of similarity, individual clusters are more clearly defined (Fig. 5). For instance, Iraqi accessions from 5 out of seven groups are admixtures with at least some portion of the genome sharing Iraqspecific alleles (Fig. 5). A new distinct group appeared in UAE accessions. Indian accessions were more like African accessions in all STRUCTURE analyses with k > 2, and Pakistani accessions clustered with Indian accessions (Figs. 4 and 5).
4. DiscussionThis study presents an intrageneric phylogeny of Phoenix derived from 52 shared chloroplast genes and whole plastomes of 29 accessions representing 14 Phoenix species. This analysis is a substantial improvement relative to prior studies of the genus phylogeny, which used very few loci and/or few species. The phylogenetic trees based on the shared plastid gene sequences and the whole plastome sequences agree in that the cultivated date palm accessions are most closely related to the male and female accessions of P. sylvestris, Phoenix cespitosa, P. atlantica, and male accessions of P. acaulis, and P. pusilla. Some recent literature considers that P. atlantica, which is a native to Cape Verde islands, may not be a separate species but rather a feral population of P. dactylifera that was naturally vectored to these isolated islands from cultivated date palms in Africa (Gros-Balthazard et al., 2017; Pintaud et al., 2010). Of the other four species in the dactylifera clade (Group 1), three species (P. sylvestris, P. acaulis, P. pusilla) are native to the Indian subcontinent, while P. cespitosa is native to the Arabian Peninsula and Somalia. The plastome-based phylogenetic trees showed marked cytonuclear discordance when compared with the nuclear gene-based (Cyp703) phylogenetic tree reported in Torres et al. (2018). The Cyp703-based phylogenetic tree suggested that P. dactylifera is more closely related to P. theophresti, P. atlantica and P. reclinata than to Phoenix sylverstris. However, both these studies agree that P. sylvestris may not be the direct progenitor of cultivated date palm. It could be that the interbreeding of more than one Phoenix species followed by natural and human selection might have resulted in the origin of P. dactylifera. Given that our chloroplast results only track maternal inheritance, one possible explanation for the different conclusions between our study and that of Torres et al. (2018) is that the female ancestor was closely related to P. sylvestris, while the male ancestor of P. dactylifera may have been a species (perhaps now extinct) that was no more closely related to P. sylvestris than it was to P. acaulis, P. cespitosa or P. pusilla.
Our microsatellite data analysis did not provide any evidence regarding the most closely-related species to domesticated P. dactylifera among the wild Phoenix species. P. canariensis, P. hanceana, P. reclinata and P. sylvestris were all placed in a separate cluster and thus were equidistant from P. dactylifera. This is in contrast to the previous finding that P. sylvestris is the most likely progenitor to the cultivated date palm (Gros-Balthazard et al., 2017). That we did not detect an unusually close relatedness of cultivated date palm with P. sylvestris could be a function of the relatively small dataset employed in our study, compared with the whole genome comparison employed earlier (Gros-Balthazard et al., 2017). One unexpected observation, however, was that Iranian accession Zamordi of P. dactylifera clustered with P. canariensis. We expect that this is caused by a mislabeling of the Iranian material, but a further investigation of this issue is warranted.
The SSR genotyping data in this current study and in several earlier studies (Chaluvadi et al., 2014; Elmeer et al., 2011; Moussouni et al., 2017) have shown that observed heterozygosity was lower than the expected heterozygosity in date palms. Expected heterozygosity increases with an increase in the number of alleles and with an even distribution of alleles. Deeper SNP marker analyses of date palm accessions also observed the higher frequency of expected then observed heterozygosity in data palm and further indicated that long runs of homozygosity (up to 500 kb) could be found within otherwise heterozygous genotypes of date palm (Hazzouri et al., 2015a). This result could be an outcome of occasional inbreeding due to farmer selection for desirable traits (Hazzouri et al., 2015a), but could also be an outcome of mitotic recombination (Rovcanin et al., 2014). These two models can be evaluated when locations of centromeres are determined on the scaffolds, because mitotic recombination is expected to yield homozygosity from the site of recombination (e.g., double strand DNA breakage repair) that extends to the end of the chromosome arm (i.e., the telomere). The fact that a high level of heterozygosity remains in most of these accessions indicates that farmer selection or natural selection can still be acting on heterozygote versus homozygote fitness even in this vegetatively propagated crop.
Though date palms are predominantly propagated vegetatively, most of the accessions used in our study as well as in other studies (Elmeer et al., 2011; Racchi et al., 2014) are highly heterozygous and rich in allelic diversity. Although commercial groves are often exclusively female, purchased pollen can exhibit genotypedependent variability in its effects on fruit size, quality and maturity, otherwise known as 'Metaxenia' (Swingle, 1928; Crawford, 1936; Nixon, 1936). Thus, chance propagation of resultant seed, and clonal propagation appear to have shaped the evolutionary dynamics of date palm even after domestication.
The SSR genotypes classified all date palm germplasm into two major groups with one group predominantly enriched with African accessions and the other group enriched with Asian accessions. This agrees with earlier studies and also suggests that date palm may have been independently domesticated in Asia and North Africa (Hazzouri et al., 2015a; Mathew et al., 2015a). The oasis agrosystem is common for date palms in the deserts of West Asia and North Africa, and tends to generate population divergence because of the often great distances between oases. Date palm domestication along the Persian Gulf has been documented (Beech and Shepherd, 2001; Hazzouri et al., 2015a), but may not have been detected in Africa because of a lack of either early written evidence or archaeobotanical studies (Terral et al., 2012).
It is particularly interesting that the Indian accessions were genetically narrow, well differentiated from all other accessions (except Pakistani accessions) and most similar to Asian accessions at k = 2. However, with the greater differentiation ability at k = 3, Indian and Pakistani accessions showed more alleles that are present predominantly in African accessions than in Asian accessions. A greater similarity of African and Pakistani accessions was also predicted in a previous study (Mathew et al., 2015). These results suggest that India and Pakistan received their date palm germplasm primarily from Africa, and not from the nearer germplasm sources in the Middle East. Whether this reflects specific trade patterns, specific cultural relationships or shared environmental demands of transplanted date palms is not clear. However, these results suggest that investigation of date palms in Somalia and Ethiopia might be particularly informative.
Date palms were first introduced into the New World by the Spanish during the colonial period. The industrial planting of date palm began in the late 1800s, mostly in the low desert areas of California and Arizona. To support this industry, the United States Department of Agriculture imported germplasm beginning in the late 1800s (Johnson, 2010). Although the main cultivars grown in the US are of North African origin, the germplasm collection includes varieties of Asian origin as well as locally developed varieties (Wright, 2016). Most of the US accessions used in our study have records regarding their donors and place of origin, but some do not. Our analyses of the USDA accessions without prior information on origin showed that they are primarily from North Africa. These USDA collections are already seeing a reverse migration, as several Arab countries undertook large-scale programs to increase date palm acreage but found a shortage of suitable offshoots. Primarily because of the absence of major pests and pathogens, California and Arizona have become highly desirable sources of offshoots (Johnson, 2010).
We observed that independent samples of genotypes with the same name, collected from different locations, usually clustered together in phylogenetic analyses. Starting from an offshoot is expected to dramatically decrease the likelihood of genetic divergence between samples of the same cultivar, and the farmer has a secondary check on accession validity because each cultivar has very distinctive fruit traits. However, differences were observed in all accessions with the same name, confirming that mutation is ongoing, especially for highly polymorphic markers like SSRs. In a few cases (e.g., for some Medjool sources), dramatic variation was observed, suggesting unintentional outcrosses. According to Devanand and Chao (2003) and Elhoumaizi et al. (2005), Medjool can be considered an ancient landrace as well as a modern cultivar. The SSR analysis described herein is an efficient and definitive technology for discerning such disparities from the expected genotypic constitution, and thus may become a tool for routine germplasm assessment and verification.
5. ConclusionsOur results show that the plastomes of cultivated date palm accessions are most closely and about equally related to the plastomes of the male and female accessions of P. sylvestris, P. caespitosa and male accessions of P. acualis, and P. pusilla. Future comparisons of Phoenix nuclear and organellar genomes, using our results as a baseline, should be able to identify the level of intercrossing between other Phoenix species and P. dactylifera, with the conflicting potential to both erode natural variation in wild species and to provide new allelic variation for domesticated date palm improvement. The date palm accessions display high genetic diversity and relatively low observed heterozygosity in the analyzed SSR loci. The accessions are genetically structured according to their geographic origin and form two main groups, African and Asian. Most of the US accessions, both with known and with unknown origins, are closely related to African genotypes. Indian and Pakistani date palms appear to have a distant African origin rather than an Asian origin, while Tunisia is unusual for its robust mixture of both African and Asian genotypes. Future studies on date palm germplasm should be targeted on providing information regarding the importance of heterozygosity versus homozygosity in particular genomic regions of the date palm genome. These analyses could indicate mechanism(s) for the origin and possible agronomic value of the homozygous regions that are consciously or unconsciously selected in breeding programs. This study also suggests future direction regarding the sources of alleles related to geographical adaptation and future breeding for improved cultivars.
Conflict of interestThe authors declare no conflict of interests.
AcknowledgmentsThis research was funded by endowment funds from the University of Georgia Giles Professorship and the Georgia Research Alliance. The authors also thank Jeff Wagner at the Georgia Genomics and Bioinformatics Core for assistance with SSR genotyping.
Appendix A. Supplementary dataSupplementary data to this article can be found online at https://doi.org/10.1016/j.pld.2018.11.005.
Abbo S., Gopher A., Lev-Yadun S., 2015. Fruit domestication in the near east. Plant Breed. Rev, 39: 325-377. |
Ahmed T.A., Al-Qaradawi A.Y., 2009. Molecular phylogeny of Qatari date palm genotypes using simple sequence repeats markers. Biotechnology, 8: 126-131. DOI:10.3923/biotech.2009.126.131 |
Akkak A., Scariot V., Marinoni D.T., Boccacci P., Beltramo C., Botta R., 2009. Development and evaluation of microsatellite markers in Phoenix dactylifera L. and their transferability to other Phoenix species. Biol. Plant, 53: 164-166. |
Al-Abdoulhadi I., Al-Ali S., Khurshid K., Al-Shryda F., Al-Jabr A., Abdallah A.B., 2011. Assessing fruit characteristics to standardize quality norms in date cultivars of Saudi Arabia. Indian J. Sci. Technol, 4: 1262-1266. |
Al-Ruqaishi I.A., Davey M., Alderson P., Mayes S., 2008. Genetic relationships and genotype tracing in date palms (Phoenix dactylifera L.) in Oman, based on microsatellite markers. Plant Genet. Resour, 6: 70-72. DOI:10.1017/S1479262108923820 |
Arabnezhad H., Bahar M., Mohammadi H.R., Latifian M., 2012. Development, characterization and use of microsatellite markers for germplasm analysis in date palm (Phoenix dactylifera L.). Sci. Hortic-Amst, 134: 150-156. DOI:10.1016/j.scienta.2011.11.032 |
Barrow S.C., 1998. A monograph of Phoenix L. (palmae:Coryphoideae). Kew Bull: 513-575. |
Beech M., Shepherd E., 2001. Archaeobotanical evidence for early date consumption on Dalma Island, United Arab Emirates. Antiquity, 75: 83. DOI:10.1017/S0003598X00052765 |
Cavalli-Sforza L.L., Edwards A.W., 1967. Phylogenetic analysis:models and estimation procedures. Evolution, 21: 550-570. DOI:10.1111/j.1558-5646.1967.tb03411.x |
Chaluvadi S.R., Khanam S., Aly M.A.M., Bennetzen J.L., 2014. Genetic diversity and population structure of native and introduced date palm (Phoenix dactylifera) germplasm in the United Arab Emirates. Trop. Plant. Biol, 7: 30-41. DOI:10.1007/s12042-014-9135-7 |
Crawford, C., 1936. The growth rate of Deglet Noor dates in metaxenia. In: Amer. Soc. Hort. Sci. Proc., pp. 51-54.
|
Devanand P., Chao C., 2003. Genetic variation within 'Medjool' and 'Deglet Noor'-date (Phoenix dactylifera L.) cultivars in California detected by fluorescent-AFLP markers. J. Hortic. Sci. Biotechnol, 78: 405-409. DOI:10.1080/14620316.2003.11511639 |
Darling A.C., Mau B., Blattner F.R., Perna N.T., 2004. Mauve:multiple alignment of conserved genomic sequence with rearrangements. Genome Res, 14: 1394-1403. DOI:10.1101/gr.2289704 |
Darling A.E., Mau B., Perna N.T., 2010. progressiveMauve:multiple genome alignment with gene gain, loss and rearrangement. 5, e11147.. PLoS One, 5: e11147. DOI:10.1371/journal.pone.0011147 |
Diez C.M., Trujillo I., Martinez-Urdiroz N., Barranco D., Rallo L., Marfil P., Gaut B.S., 2015. Olive domestication and diversification in the mediterranean basin. New Phytol, 206: 436-447. DOI:10.1111/nph.13181 |
Earl D.A., 2012. STRUCTURE HARVESTER:a website and program for visualizing STRUCTURE output and implementing the Evanno method. Conserv. Genet. Resour, 4: 359-361. DOI:10.1007/s12686-011-9548-7 |
El-Juhany L.I., 2010. Degradation of date palm trees and date production in Arab countries:causes and potential rehabilitation. Aust. J. Basic Appl. Sci, 4: 3998-4010. |
Elhoumaizi M.A., Devanand P.S., Fang J., Chao C.-C.T., 2005. Confirmation of Medjool'date as a landrace variety through genetic analysis of 'medjool' accessions in Morocco. J. Am. Soc. Hortic. Sci, 131: 403-407. |
Elmeer K., Mattat I., 2015. Genetic diversity of Qatari date palm using SSR markers. Genet. Mol. Res, 14: 1624-1635. DOI:10.4238/2015.March.6.9 |
Elmeer K., Sarwath H., Malek J., Baum M., Hamwieh A., 2011. New microsatellite markers for assessment of genetic diversity in date palm (Phoenix dactylifera L.). Biotech, 1: 91-97. |
Elshibli S., Korpelainen H., 2008. Microsatellite markers reveal high genetic diversity in date palm (Phoenix dactylifera L.) germplasm from Sudan. Genetica, 134: 251-260. DOI:10.1007/s10709-007-9232-8 |
Erskine, W., Moustafa, A.T., Osman, A.E., Lashine, Z., Nejatian, A., Badawi, T., Ragy, S.M., 2004. Date Palm in the GCC Countries of the Arabian Peninsula. In: Proc. Regional Workshop on Date Palm Development in the Arabian Peninsula, Abu Dhabi, UAE.
|
Evanno G., Regnaut S., Goudet J., 2005. Detecting the number of clusters of individuals using the software STRUCTURE:a simulation study. Mol. Ecol, 14: 2611-2620. DOI:10.1111/j.1365-294X.2005.02553.x |
Excoffier L., Smouse P.E., Quattro J.M., 1992. Analysis of molecular variance inferred from metric distances among DNA haplotypes:application to human mitochondrial DNA restriction data. Genetics, 131: 479-491. |
Frailey D.C., Chaluvadi S.R., Vaughn J.N., Coatney C.G., Bennetzen J.L., 2018. Gene loss and genome rearrangement in the plastids of five Hemiparasites in the family Orobanchaceae. BMC Plant Biol, 18: 30. DOI:10.1186/s12870-018-1249-x |
Govaerts, R., Dransfield, J., 2005. World Checklist of Palms. Royal Botanic Gardens, Kew: Kew, UK, p. 235.
|
Gros-Balthazard M., Galimberti M., Kousathanas A., Newton C., Ivorra S., Paradis L., Vigouroux Y., Carter R., Tengberg M., Battesti V., 2017. The discovery of wild date palms in Oman reveals a complex domestication history involving centers in the Middle East and Africa. Curr. Biol, 27: 2211-2218. DOI:10.1016/j.cub.2017.06.045 |
Hamza H., Elbekkay M., Ben Abederrahim M.A., Ali A.F., 2011. Molecular and morphological analyses of date palm (Phoenix dactylifera L.) subpopulations in southern Tunisia. Spanish J. Agric. Res, 9: 484-493. DOI:10.5424/sjar/20110902-271-10 |
Hamza H., Benabderrahim M.A., Elbekkay M., Ferdaous G., Triki T., Ferchichi A., 2012. Investigation of genetic variation in Tunisian date palm (Phoenix dactylifera L.) cultivars using ISSR marker systems and their relation with fruit characteristics. Turk. J. Biol, 36: 449-458. |
Hazzouri K.M., Flowers J.M., Visser H.J., Khierallah H.S.M., Rosas U., Pham G.M., Meyer R.S., Johansen C.K., Fresquez Z.A., Masmoudi K., Haider N., El Kadri N., Idaghdour Y., Malek J.A., Thirkhill D., Markhand G.S., Krueger R.R., Zaid A., Purugganan M.D., 2015. Whole genome re-sequencing of date palms yields insights into diversification of a fruit tree crop. Nat. Commun, 6: 8824. DOI:10.1038/ncomms9824 |
Jakobsson M., Rosenberg N.A., 2007. CLUMPP:a cluster matching and permutation program for dealing with label switching and multimodality in analysis of population structure. Bioinformatics, 23: 1801-1806. DOI:10.1093/bioinformatics/btm233 |
Johnson, D., 2010. Worldwide Dispersal of the Date Palm from its Homeland. In: IV International Date Palm Conference, vol. 882, pp. 369-375.
|
Jubrael J.M.S., Udupa S.M., Baum M., 2005. Assessment of AFLP-based genetic relationships among date palm (Phoenix dactylifera L.) varieties of Iraq. J. Am. Soc. Hortic. Sci, 130: 442-447. DOI:10.21273/JASHS.130.3.442 |
Larkin M.A., Blackshields G., Brown N., Chenna R., Mcgettigan P.A., Mcwilliam H., Valentin F., Wallace I.M., Wilm A., Lopez R., 2007. Clustal W and clustal X version 2., 0. Bioinformatics, 23: 2947-2948. DOI:10.1093/bioinformatics/btm404 |
Mathew L.S., Seidel M.A., George B., Mathew S., Spannagl M., Haberer G., Torres M.F., Al-Dous E.K., Al-Azwani E.K., Diboun I., 2015. A genome-wide survey of date palm cultivars supports two major subpopulations in Phoenix dactylifera. G3:Genes. Genetics, 5: 1429-1438. |
Minch, E., Ruiz-Linares, A., Goldstein, D., Feldman, M., Cavalli-Sforza, L., 1995. Microsat (Version 1.4 D): a Computer Program for Calculating Various Statistics on Microsatellite Allele Data. www.lotka.stanford.edu/microsat.html.
|
Morrell P.L., Buckler E.S., Ross-Ibarra J., 2012. Crop genomics:advances and applications. Nat. Rev. Genet, 13: 85-96. DOI:10.1038/nrg3097 |
Moussouni S., Pintaud J.-C., Vigouroux Y., Bouguedoura N., 2017. Diversity of Algerian oases date palm (Phoenix dactylifera L., Arecaceae):heterozygote excess and cryptic structure suggest farmer management had a major impact on diversity. PLoS One, 12: e0175232. DOI:10.1371/journal.pone.0175232 |
Nei M., Chakraborty R., 1973. Genetic distance and electrophoretic identity of proteins between taxa. J. Mol. Evol, 2: 323-328. DOI:10.1007/BF01654100 |
Nixon R.W., 1936. Metaxenia and interspecific pollinations in Phoenix. Proc. Am. Soc. Hortic. Sci, 33: 21-26. |
Peakall R., Smouse P.E., 2012. GenAlEx 6., 5:genetic analysis in Excel. Population genetic software for teaching and research-an update. Bioinformatics, 28: 2537-2539. DOI:10.1093/bioinformatics/bts460 |
Pintaud, J.-C., Zehdi, S., Couvreur, T., Barrow, S., Henderson, S., Aberlenc-Bertossi, F., Tregear, J., Billotte, N., 2010. Species delimitation in the genus Phoenix (Arecaceae) based on SSR markers, with emphasis on the identity of the date palm(Phoenix dactylifera L.). In: Diversity, Phylogeny, and Evolution in the Monocotyledons. Arhus University Press, Denmark, pp. 267-286.
|
Pintaud, J.-C., Ludeña, B., Aberlenc-Bertossi, F., Zehdi, S., Gros-Balthazard, M., Ivorra, S., Terral, J.-F., Newton, C., Tengberg, M., Abdoulkader, S., 1994. Biogeography of the date palm (Phoenix dactylifera L., Arecaceae): insights on the origin and on the structure of modern diversity. Acta Hortic. 2013, 9-38.
|
Pritchard J.K., Stephens M., Donnelly P., 2000. Inference of population structure using multilocus genotype data. Genetics, 155: 945-959. |
Racchi M., Bove A., Turchi A., Bashir G., Battaglia M., Camussi A., 2014. Genetic characterization of Libyan date palm resources by microsatellite markers. Biotech, 4: 21-32. |
Ronquist F., Teslenko M., Van Der Mark P., Ayres D.L., Darling A., Höhna S., Larget B., Liu L., Suchard M.A., Huelsenbeck J.P., 2012. MrBayes 3. 2:efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol, 61: 539-542. DOI:10.1093/sysbio/sys029 |
Rousset F., 2008. genepop'007:a complete re-implementation of the genepop software for Windows and Linux. Mol. Ecol. Resour, 8: 103-106. DOI:10.1111/j.1471-8286.2007.01931.x |
Rovcanin B., Ivanovski I., Djuric O., Nikolic D., Petrovic J., Ivanovski P., 2014. Mitotic crossover-an evolutionary rudiment which promotes carcinogenesis of colorectal carcinoma. World J. Gastroenterol, 20: 12522. DOI:10.3748/wjg.v20.i35.12522 |
Sedra, M., 2010. Evaluation of soil receptivity of date palm groves in Arab countries to Fusarium oxysporum f. sp. albedinis, causal agent of bayoud disease of date palm. In: IV International Date Palm Conference, vol. 882, pp. 515-525.
|
Stich B., Melchinger A.E., Frisch M., Maurer H.P., Heckenberger M., Reif J.C., 2005. Linkage disequilibrium in European elite maize germplasm investigated with SSRs. Theor. Appl. Genet, 111: 723-730. DOI:10.1007/s00122-005-2057-x |
Swingle W.T., 1928. Metaxenia in the date palm:possibly a hormone action by the embryo or endosperm. J. Hered, 19: 257-268. DOI:10.1093/oxfordjournals.jhered.a102996 |
Tengberg M., 2012. Beginnings and early history of date palm garden cultivation in the Middle East. J. Arid Environ, 86: 139-147. DOI:10.1016/j.jaridenv.2011.11.022 |
Terral J.F., Newton C., Ivorra S., Gros-Balthazard M., de Morais C.T., Picq S., Tengberg M., Pintaud J.C., 2012. Insights into the historical biogeography of the date palm (Phoenix dactylifera L.) using geometric morphometry of modern and ancient seeds. J. Biogeogr, 39: 929-941. DOI:10.1111/j.1365-2699.2011.02649.x |
Torres M., Mathew L., Ahmed I., Al-Azwani K. I., Krueger R., Rivera-Nuñez D. A., Mohamoud Y., Clark G. A., Suhre K., Malek A. J., 2018. Genus-wide sequencing supports a two-locus model for sex-determination in Phoenix. Nat. Commun, 9: 3969. DOI:10.1038/s41467-018-06375-y |
Vaughn J.N., Chaluvadi S.R., Rangan L., Bennetzen J.L., 2014. Whole plastome sequences from five ginger species facilitate marker development and define limits to barcode methodology. PLoS One, 9: e108581. DOI:10.1371/journal.pone.0108581 |
Wagner, H., Sefc, K., 1999. IDENTITY 1.0 Centre for Applied Genetics. University of Agricultural Sciences, Vienna. http:/boku.ac.at/zag/forsgh/identity.htm.
|
Wales N., Blackman B.K., 2017. Plant domestication:wild date palms illuminate a crop's sticky origins. Curr. Biol, 27: R702-R704. DOI:10.1016/j.cub.2017.05.070 |
Wright G.C., 2016. The commercial date industry in the United States and Mexico. Hortscience, 51: 1333-1338. DOI:10.21273/HORTSCI11043-16 |
Wright S., 1950. Genetical structure of populations. Nature, 166: 247-249. DOI:10.1038/166247a0 |
Wyman S.K., Jansen R.K., Boore J.L., 2004. Automatic annotation of organellar genomes with DOGMA. Bioinformatics, 20: 3252-3255. DOI:10.1093/bioinformatics/bth352 |
Zehdi-Azouzi S., Cherif E., Moussouni S., Gros-Balthazard M., Naqvi S.A., Ludena B., Castillo K., Chabrillange N., Bouguedoura N., Bennaceur M., SiDehbi F., Abdoulkader S., Daher A., Terral J.F., Santoni S., Ballardini M., Mercuri A., Ben Salah M., Kadri K., Othmani A., Littardi C., Salhi-Hannachi A., Pintaud J.C., Aberlenc-Bertoss F., 2015. Genetic structure of the date palm(Phoenix dactylifera) in the Old World reveals a strong differentiation between eastern and western populations. Ann. Bot-Lond, 116: 847-847. DOI:10.1093/aob/mcv132 |
Zhao Y., Williams R., Prakash C.S., He G., 2012. Identification and characterization of gene-based SSR markers in date palm (Phoenix dactylifera L.). BMC Plant Biol, 12: 237. DOI:10.1186/1471-2229-12-237 |



