b. University of Chinese Academy of Sciences, Beijing, 100049, China;
c. CAS Key Laboratory for Plant Diversity and Biogeography of East Asia, Kunming Institute of Botany, Chinese Academy of Sciences, Kunming, Yunnan, 650201, China
Myripnois Bunge (Asteraceae) is a monotypic genus endemic to China. Myripnois dioica Bunge can only be found in northern China and predominantly grows in the shadow slopes of mountains. It is a rare shrub in the herb-dominated daisy family. Owing to its purple and white hermaphrodite flowers, M. dioica is thought to be a potential ornamental plant (Xie and Zhang, 2012).
Currently, the phylogenetic position of Myripnois is controversial. Early studies suggested that Myripnois, Ainsliaea DC., Pertya Sch. Bip., and the Japanese endemic Macroclinidium Maxim. comprise the tribe Pertyeae (Pertyoideae) (Funk et al., 2009; Panero and Funk, 2002). Flora of China, however, places Myripnois together with eight other Asteraceae genera (Adenocaulon Hook., Ainsliaea, Gerbera L., Leibnitzia Cass., Leucomeris D. Don, Nouelia Franch., Pertya and Piloselloides (Less.) C.Jeffrey ex Cufod.) in the tribe Mutisieae s.l. (Gao and Nicholas, 2011). Leucomeris and Nouelia have since been moved to Wunderlichieae (Wunderlichioideae), whereas Adenocaulon, Gerbera (including Piloselloides) and Leibnitzia have remained in Mutisieae s.str. (Mutisioideae) (Panero et al., 2014). Thus, the tribe Pertyeae may include three genera (Myripnois, Ainsliaea, and Pertya). More recently, phylogenetic studies have revealed that Myripnois is more closely related to Peryta than to Ainsliaea (Fu et al., 2016). Furthermore, Freire (2017) suggested that Myripnois should be included in Peryta; however, this treatment has not been adopted broadly.
The megafamily Asteraceae was initially divided into 12 subfamilies (Panero and Funk, 2002, 2008): Asteroideae, Barnadesioideae, Carduoideae, Cichorioideae, Corymbioideae, Gochnatioideae, Gymnarrhenoideae, Hecastocleidoideae, Mutisioideae, Pertyoideae, Stifftioideae and Wunderlichioideae. Molecular evidence later recovered the new subfamily Famatinanthoideae (Panero et al., 2014). Many phylogenetic studies have investigated taxonomic relationships within Asteraceae at both tribe and genus levels (e.g., Fu et al., 2016; Nie et al., 2015; Vargas et al., 2017). Therefore, even though these studies suffer from relatively sparse sampling, the backbone of Asteraceae at subfamily level has been generally constructed (i.e. Fu et al., 2016; Funk et al., 2009; Panero and Crozier, 2016; Panero et al., 2014). However, rapid radiation and rampant polyploidy events, which frequently occur in Asteraceae (Barker et al., 2008, 2016; Huang et al., 2016), have likely stymied efforts to resolve phylogenetic relationships within subfamilies (Mandel et al., 2017). Genomic data, including plastomes, may help clarify these phylogenetic relationships.
Angiosperm plastomes have proven effective in resolving relationships at different taxonomic levels via variation of sequences (e.g., Jansen et al., 2007; Li et al., 2019; Ruhfel et al., 2014) and structure (e.g., Sun et al., 2017, 2018). To date, 152 Asteraceae plastomes have been sequenced, but only three subfamilies (Asteroideae, Cichorioideae and Carduoideae) are covered. Previous studies (e.g., Jansen and Palmer, 1987; Kim et al., 2005) revealed that a ~20-kb inversion and a ~ 3-kb inversion are shared by most Asteraceae plastomes. In addition, the gene content of the inverted repeat regions of Asteraceae plastomes have been shown to vary (e.g., Vargas et al., 2017; Wang et al., 2017; Wei et al., 2015; Zhang et al., 2014; Zhang et al., 2017).
In the present study, our goal was to examine both plastome evolution and elucidate the phylogenetic relationships within Asteraceae. For this purpose, we first sequenced and characterized the plastome of M. dioica; we then compared the M. dioica plastome to those of 77 Asteraceae species. To elucidate the phylogenetic relationships within Asteraceae, we used the plastomes of 78 Asteraceae representatives. Our analyses identified multiple simple sequence repeats (SSRs) within M. dioica and hotspot regions of sequence variation within Asteraceae, both of which may contribute to future studies on population genetics.
2. Materials and methods 2.1. Plant materials, sequencing, and assemblyFresh leaves of M. dioica from Taibai Mountain (34.1864° N, 108.0077° E), Shaanxi Province (China), were sampled and dried with silica gel. A voucher specimen was deposited in the Herbarium of the Wuhan Botanical Garden, Chinese Academy of Sciences (HIB 20170527). Chloroplast genomic DNA was extracted from approximately 5 mg of silica-dried leaf tissue following the methods of Shi et al. (2012). The quality and concentration of DNA were assessed using agarose gel electrophoresis and an Agilent BioAnalyzer 2100 (Agilent Technologies, California, USA). Purified DNA was used to generate short-insert (500 bp) paired-end sequencing libraries according to the Illumina standard protocol, and sequencing was performed by Beijing Genomics Institute (Wuhan, Hubei, China). Raw sequence reads produced by Illumina paired-end sequencing were filtered using NGS QC Tool Kit (Patel and Jain, 2012) by removing adapter sequences and low-quality reads with Q value B20. The reads of high quality were subsequently assembled into contigs with a minimum length of 1000 bp using CLC Genomics Workbench 9 with the Kmer = 60 to get maximal N50 value (Girard et al., 2011).
2.2. Genome annotation and comparative plastome analysisInitial gene location of the complete plastome was determined with Dual Organellar GenoMe Annotator (DOGMA) (Wyman et al., 2004). Then, annotations for the start and stop codons and intron/ exon boundaries were manually adjusted using ten published Asteraceae plastomes. tRNA genes were verified with tRNAscan-SE (Lowe and Eddy, 1997). A circular physical genome map was drawn using the GenomeVx (Conant and Wolfe, 2008) and subsequently edited manually. The structure, gene content, and order of 78 Asteraceae plastomes were compared (Table S1). To explore the structural evolution among 78 Asteraceae plastomes, we listed genes within the IR/SC boundaries and mapped them on the phylogenomic tree. We also summarized the plastome size and life form of these 78 Asteraceae species and did Spearman's correlation test using R (R Core Team, 2013). The sequence identity plots among 11 plastomes representing all available tribes within Asteraceae were visualized using mVISTA in shuffle-LAGAN mode (Frazer et al., 2004) with Artemisia frigida Willd. as the reference. In order to screen variable characters among plastomes within Asteraceae, the average number of nucleotide differences was determined to analyze nucleotide diversity (Pi) using DnaSP v5.0 (Rozas et al., 2003).
2.3. SSR identificationPrevious studies showed that repetitive sequences are useful in studying plastome evolution and rearrangement (George et al., 2015; Govindaraj et al., 2015). Due to the high polymorphism and abundance, simple sequence repeats (SSRs) have been extensively used as molecular markers for phylogenetic research, population genetics, and evolutionary studies (Wheeler et al., 2014; Xue et al., 2012). SSRs were identified from the M. dioica plastome with MISA (Thiel et al., 2003) by setting the minimum number of repeats to 10, 5, and 4 for mono-, di-, trinucleotide repeats and 3 for tetra-, penta-, hexa-, hepta-, octa-, nona-, and decanucleotide repeats. In general, large repeated elements comprise four types: direct (forward), inverted (palindromic), complement, and reverse repeats. We further identified these four types of repeat sequences in the M. dioica plastome using the online REPuter software with a minimum repeat size of 20 bp and sequence identity greater than 90% (Kurtz et al., 2001).
2.4. Phylogenetic analyses within AsteraceaePlastomes of 81 species, including all available genera representatives of Asteraceae and three outgroups (Hanabusaya asiatica (Nakai) Nakai, Adenophora remotiflora (Siebold & Zucc.) Miq. and Codonopsis minima Nakai) (Table S1) (Cheon et al., 2017; Cheon and Yoo, 2016; Kim et al., 2016), were included for phylogenetic analyses. The sequences of 79 shared protein-coding genes were extracted from all 81 plastomes and aligned using MAFFT v. 7 software (Katoh and Toh, 2010). Both partitioned and unpartitioned Bayesian inference (BI) were conducted using MrBayes v.3.2 (Ronquist et al., 2012). The general time-reversible (GTR) substitution model determined by Modeltest 3.7 (Posada and Crandall, 1998) was selected as the optimal model. The best-fit partition scheme containing 14 partitions (maximum likelihood score [ln L] = -314725.9017, Bayesian information criterion [BIC] = 632896.9804) was determined by PartitionFinder version 1.1.1 (Lanfear et al., 2012). BI analysis was set to run for 2, 000, 000 generations and sampled every 100 generations with the first 25% of the trees regarded as burn-in (Ronquist et al., 2012).
3. Results 3.1. Plastome structure of MyripnoisThe sequencing generated a total of 5, 062, 401 clean reads (Table 1). De novo and reference-guided assembly finally produced a single scaffold comprising the entire plastome sequence. The plastome displayed a typical quadripartite structure, consisting of a pair of inverted repeat (IR) regions (25, 212 bp) separated by a large single-copy (LSC) region (84, 271 bp) and a small single-copy (SSC) region (18, 437 bp) (Fig. 1). The plastome contains 113 unique genes, including 79 protein-coding genes, 30 tRNA genes, and 4 rRNA genes (Table S2). The average G/C content is 38.7%. Additionally, 14 genes possess one intron, and three genes (rps12, clpP, ycf3) possess two introns (Table S2). A 20-kb inversion, including 16 genes from trnS-GCU to trnE-UUG, was observed in the LSC region. One additional ~3-kb inversion (located between trnC-GCA and trnE-UUC) was also detected within the 20-kb inversion (Fig. 1). Thirteen SSRs ≥10 bp were identified, including 12 mono- and one trinucleotide SSRs. The highest portion of mononucleotide SSRs were A (46.15%) and T (38.46%) repeat units (Table S3). We identified 58 repeats larger than 20 bp and with a sequence identity >90%, including 27 direct, nine reverse, two complement, and 20 palindromic repeats (Table S4). The lengths of repeats range from 21 to 48 bp. Finally, we found the repeats of M. dioica plastome are mainly located in intergenic spacer (77.59%), while a minority are located in introns or gene coding regions (22.41%) (Table S4).
| Plastome characteristics | |
| Raw read number | 5, 062, 401 |
| Total read length (bp) | 759, 360, 150 |
| Mapped read number | 1, 817, 562 |
| Plastome coverage (x) | 237 |
| Total length (bp) | 153, 132 |
| LSC length (bp) | 84, 271 |
| SSC length (bp) | 18, 437 |
| IR length (bp) | 25, 212 |
| G/C content (%) | 37.6 |
| Number of genes in the IR | 18 |
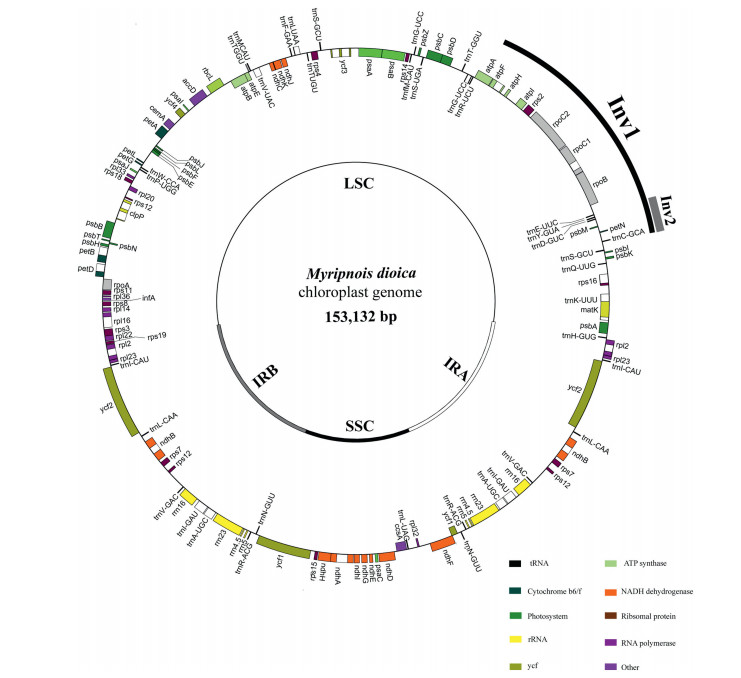
|
| Fig. 1 Physical map of the Myripnois dioica plastome. |
All Asteraceae plastomes are composed of two IR regions (23, 595-26, 916 bp) separated by the LSC (81, 998-85, 747 bp) and SSC (13, 325-19, 324 bp) regions (Table S1). The overall G/C content ranged from 37.10% (in Anaphalis sinica Hance) to 37.80% (in multiple plastomes).
Plastome and plastome region sizes vary among Asteraceae species. Overall plastome size in Asteraceae species ranges from 149, 473 bp (Aster spathulifolius Maxim.) to 171, 724 bp (Baccharis genistelloides (Lam.) Pers.). The LSC region ranges from 81, 998 bp (A. spathulifolius) to 85, 311 bp (Pericallis hybrida B.Nord.), whereas the IR region ranges from 23, 595 bp (P. hybrida) to 26, 916 bp (Dendrosenecio johnstonii (Oliv.) B.Nord.). Finally, the SSC region ranges from 13, 325 bp (D. johnstonii) to 19, 324 bp (Leontopodium leiolepis Nakai). For the 78 Asteraceae species examined in this study, plastome size is not significantly correlated with life form (P = 0.14, Table S1; Fig. S1).
In addition, the boundary regions of Asteraceae plastomes vary. For instance, in most Asteraceae plastomes the ycf1 gene is located at the SSC/IR junctions; however, in Achyrachaena mollis Schauer., Chrysanthemum × morifolium Ramat. and Artemisia argyi H. Lev. & Vaniot, this gene is not present at the SSC/IR junction. Furthermore, we detected significant instability within Asteraceae at the LSC/IR boundary (Fig. 2).
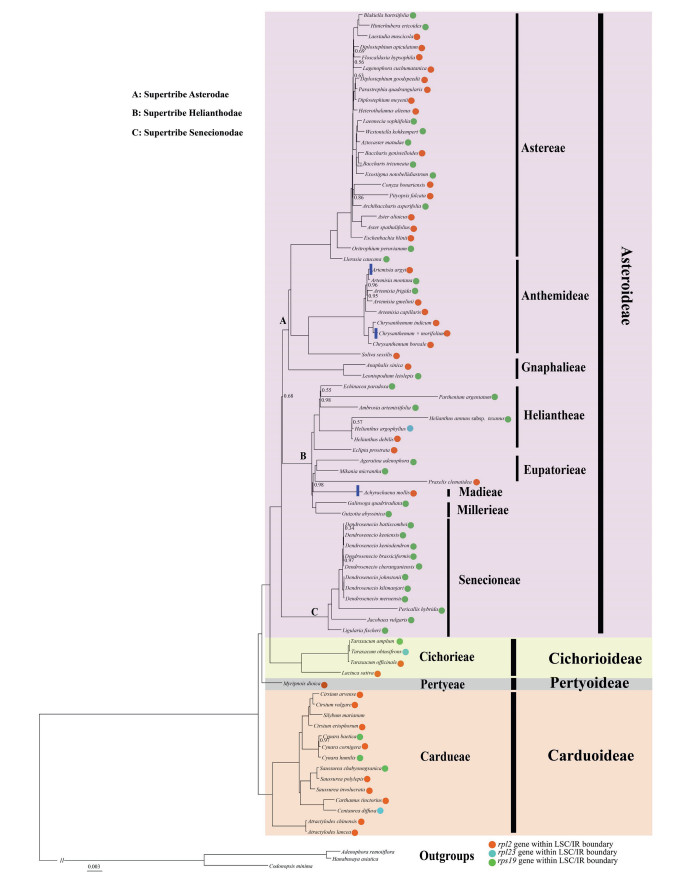
|
| Fig. 2 The Bayesian inference tree of Asteraceae based on the 14-partition analysis of 79 plastid protein coding genes. Only the branches with posterior probabilities support less than 1.00 are labeled with corresponding values. Branches with or without blue rectangle represent the ycf1 gene within or outside of the IR/SSC boundary. |
Eleven Asteraceae tribes show a high degree of genetic heterogeneity (Fig. 3). The single-copy regions (LSC and SSC) are more genetically differentiated than the IR region (Fig. 3). The lowest and highest sequence divergence of protein-coding gene is 0.26% and 6.41% in psbL and rps15, respectively (Fig. 4 and Table S5).
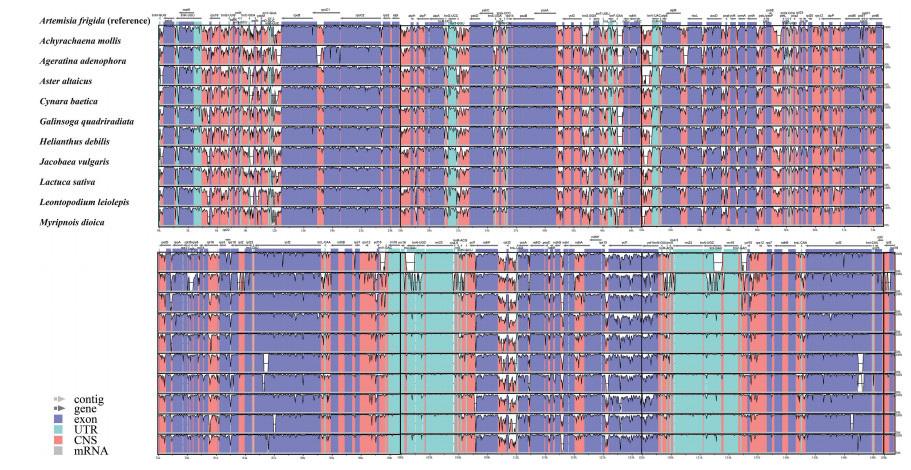
|
| Fig. 3 Structural alignment among 11 Asteraceae plastomes. VISTA-based identity plots show sequence identity among Asteraceae plastomes; sequence identity is shown as a percentage between 50 and 100% on the y-axis with Artemisia frigida as reference. The arrows indicate the direction of transcription. |
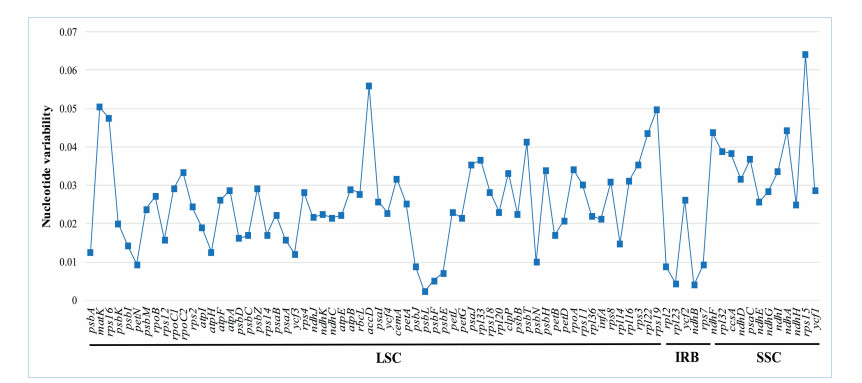
|
| Fig. 4 Plot of nucleotide variability values among Asteraceae plastomes. |
The 79-gene, 81-taxon alignment was 79, 336 bp in length and used for following phylogenomic analyses. Both partitioned and unpartitioned Bayesian inference analyses were performed, and finally yielded an identical topology (Fig. 2, Table S6). Most clades were strongly supported with posterior probabilities (PP) of 1.00 (Fig. 2). Three subfamilies (Asteroideae, Cichorioideae and Carduoideae) were resolved as monophyletic groups (Fig. 2). Pertyoideae (represented by M. dioica) was sister to Asteroideae + Cichorioideae; Carduoideae further showed a sister relationship with Pertyoideae + Asteroideae + Cichorioideae. Within Asteroideae, the basal clade Senecionodae was sister to Helianthodae + Asterodae with strong support (PP = 1.00). Diplostephium (Astereae) did not form a monophyletic clade, with three Diplostephium species scattered in two different clades. Within Anthemideae, two monophyletic clades (corresponding to five Artemisia and three Chrysanthemum species) were detected; these two clades together showed a sister relationship with Soliva sessilis Ruiz & Pav. (PP = 1.00). In this study, the monophyletic Heliantheae alliance (Heliantheae + Eupatorieae + Madieae + Millerieae) is well supported (PP = 1.00), and is composed of two subclades: Heliantheae and Eupatorieae + Madieae + Millerieae. Within Carduoideae, our analysis embedded Silybum marianum Gaertn. in the Cirsium clade.
4. Discussion 4.1. Plastome structure of Myripnois and comparison within AsteraceaeTo date, more than 150 Asteraceae plastomes (belonging to 50 genera, 11 tribes and three subfamilies) have been sequenced. However, to the best of our knowledge, no previous work has comprehensively compared these plastomes. We found that all known Asteraceae plastomes, including our newly sequenced M. dioica plastome, share two large inversions (i.e. ~ 20-kb and ~ 3-kb) in the LSC region. In previous studies, large inversions have also been reported to characterize a series of other lineages, e.g., the ~39-kb and ~24-kb inversions in Ranunculaceae (Johansson, 1999), the ~50-kb inversion in Onagraceae (Hupfer et al., 2000) and the ~49-kb and ~3.5-kb inversions in Circaeasteraceae (Sun et al., 2017). Previous studies have suggested that these large inversions may be caused by short inverted repeats (sIRs) (e.g., Chumley et al., 2006; Johansson, 1999; Hupfer et al., 2000). However, short inverted repeats are not found in M. dioica.
Among the 78 Asteraceae species in the current study, 54 are herbs, seven are shrubs, nine are trees, and eight belong to mixed growth forms. Previous studies have suggested a significant correlation between genome size and growth form, in which smaller genome sizes are correlated with trees/shrubs rather than with herbs. Some researchers have interpreted this correlation between small genome size and tree/shrub life forms to mean that there is an adaptive significance for trees/shrubs with dense stomata to have smaller genomes (Beaulieu et al., 2008; Smith, 2017). In our study, genome size and life form were not strongly correlated, which may be explained by the fact that the sizes of all Asteraceae plastomes examined were similar (Table S1).
The borders of IR/LSC and IR/SSC are relatively conserved among angiosperm plastomes, mostly positioned within rps19 and ycf1, respectively (Downie and Jansen, 2015; Raubeson et al., 2007; Shinozaki et al., 1986). Among Asteraceae species, IR gene content differs (Fig. 2; Table S1), mainly due to the instability found at the LSC/IR boundary. Specifically, when we examined the plastome sequences of 78 Asteraceae species, we found three types of LSC/IR boundaries, which differed in gene number by 1-3 genes (Fig. 2). This variation is mainly caused by different degrees of expansion of the IR into the LSC region (Goulding et al., 1996). Significant expansions have been reported in other families, such as the extreme 50-kb expansion found in Pelargonium × hortorum L.H. Bailey (Geraniaceae) (Chumley et al., 2006), the 12-kb expansion in Nicotiana acuminata (Graham) Hook.
(Solanaceae) (Goulding et al., 1996), and the 4-kb expansion in Jasminum nudiflorum Lindl. (Oleaceae) (Lee et al., 2007).
4.2. Phylogenomic analyses within AsteraceaeAlthough only 4 out of 13 Asteraceae subfamilies were sampled in the current study, our phylogenomic analyses showed high support values for most nodes (Fig. 2). The phylogenetic relationships among the four subfamilies are congruent with previous studies (e.g., Fu et al., 2016; Funk et al., 2009; Huang et al., 2016; Mandel et al., 2017; Panero and Crozier, 2016; Panero and Funk, 2002, 2008; Panero et al., 2014). Furthermore, our analyses characterized all sampled Asteraceae tribes as monophyletic. Asteroideae is the largest subfamily in Asteraceae, within which three groups corresponding to three supertribes (Senecionodae, Helianthodae and Asterodae) were identified by Panero et al. (2014), and our results confirm this circumscription. We also identified the sister relationship between Senecionodae and Helianthodae + Asterodae with strong support (PP = 1.00). This finding is slightly incongruent with that of Panero et al. (2014), who found that Helianthodae was sister to the basal clade of Senecionodae + Asterodae. This disagreement may have resulted from differences in taxon and molecular marker sampling. Furthermore, the relationship between Helianthodae and Asterodae, which was moderately supported in our analyses, requires further validation. In our phylogenomic framework the entangled Heliantheae alliance (Heliantheae + Eupatorieae + Madieae + Millerieae) was strongly supported as a monophyletic clade, which is in accordance with the results of Fu et al. (2016). Taken together with previous findings (Fu et al., 2016; Panero et al., 2014), our results indicate that there is a close relationship between Pertyoideae and Cichorioideae + Asteroideae. In contrast to previous studies (Vallès et al., 2003; Oberprieler et al., 2007), our tree supports a monophyletic grouping for five Artemisia.
Additionally, in our phylogenetic analyses, variation at the SC/IR border characterizes different clades within Asteraceae; most genera (e.g., Dendrosenecio (Hauman ex Hedberg) B. Nord., Aster L. and Chrysanthemum L.) are characterized by identical LSC/IR boundaries (Fig. 2). This homology between phylogeny and plastome structure has also been detected in many other genera, e.g., Toona (Endl.) M.Roem. (Meliaceae), Spondias L. (Anacardiaceae), Zanthoxylum L. (Rutaceae) and Acer L. (Sapindaceae) (Lin et al., 2018). Thus, the regions of the plastome we have identified in this study that have high divergence (e.g., SC/IR boundaries and SSRs) represent candidate DNA barcodes that can be used to assess phylogenetic relationships and interspecific divergence in Asteraceae (Tables S3, S4 and S5).
5. ConclusionIn present study, we sequenced and characterized the plastome of M. dioica and examined the structural variation among different Asteraceae species. The plastome of M. dioica has a typical land plant plastome structure and is highly similar to that of other Asteraceae species. Plastome variation among Asteraceae varies mainly at the SC/IR boundary. Although plastome sequences from nine subfamilies are required for further analyses, the phylogenetic relationships among and within four Asteraceae subfamilies are relatively robustly supported. Our research provides new genomic resources for Asteraceae, and will facilitate studies on the genetic conservation of M. dioica. Plastome evolution and phylogenomics require additional investigation, especially for representatives of subfamilies distributed in South America and southern Africa, which have been proposed as possible sites for the origin of Asteraceae.
Conflict of interestThe authors declared that they have no conflict of interest.
AcknowledgementsThis study was supported by the Strategic Priority Research Program of Chinese Academy of Sciences (XDA20050203), the National Key R&D Program of China (2017YFC0505200), the Major Program of the National Natural Science Foundation of China (31590823), and the National Natural Science Foundation of China (31570213).
Appendix A. Supplementary dataSupplementary data to this article can be found online at https://doi.org/10.1016/j.pld.2019.07.003.
Barker M.S., Kane N.C., Kozik A., Michelmore R.W., Matvienko M., Kozik K., et al, 2008. Multiple paleopolyploidizations during the evolution of the Asteraceae reveal parallel patterns of duplicate gene retention after millions of years. Mol. Biol. Evol, 25: 2445-2455. DOI:10.1093/molbev/msn187 |
Barker M.S., Li Z., Kidder T.I., Reardon C.R., Lai Z., Oliveira L.O., et al, 2016. Most Compositae (Asteraceae) are descendants of a paleohexaploid and all share a paleotetraploid ancestor with the Calyceraceae. Am. J. Bot, 103: 1203-1211. DOI:10.3732/ajb.1600113 |
Beaulieu J.M., Leitch I.J., Patel S., Pendharkar A., Knight C.A., 2008. Genome size is a strong predictor of cell size and stomatal density in angiosperms. New Phytol, 179: 975-986. DOI:10.1111/j.1469-8137.2008.02528.x |
Cheon K.S., Kim K.A., Han J.S., Yoo K.O., 2017. The complete chloroplast genome sequence of Codonopsis minima (Campanulaceae), an endemic to Korea. Conserv. Genet. Res, 9: 541-543. DOI:10.1007/s12686-017-0718-0 |
Cheon K.S., Yoo K.O., 2016. Complete chloroplast genome sequence of Hanabusaya asiatica (Campanulaceae), an endemic genus to Korea. Mitochondrial DNA Part A, 27: 1629-1631. DOI:10.3109/19401736.2014.958702 |
Chumley T.W., Palmer J.D., Mower J.P., Fourcade H.M., Calie P.J., Boore J.L., et al, 2006. The complete chloroplast genome sequence of Pelargonium×hortorum:organization and evolution of the largest and most highly rearranged chloroplast genome of land plants. Mol. Biol. Evol, 23: 2175-2190. DOI:10.1371/journal.pone.0143792 |
Conant G.C., Wolfe K.H., 2008. GenomeVx:simple web-based creation of editable circular chromosome maps. Bioinformatics, 24: 861-862. DOI:10.1093/bioinformatics/btm598 |
Downie S.R., Jansen R.K., 2015. A comparative analysis of whole plastid genomes from the Apiales:expansion and contraction of the inverted repeat, mitochondrial to plastid transfer of DNA, and identification of highly divergent noncoding regions. Syst. Bot, 40: 336-351. DOI:10.1600/036364415X686620 |
Frazer K.A., Pachter L., Poliakov A., Rubin E.M., Dubchak I., 2004. VISTA:computational tools for comparative genomics. Nucleic Acids Res, 32: 273-279. DOI:10.1093/nar/gkh458 |
Fu Z.X., Jiao B.H., Nie B., Zhang G.J., Gao T.G., 2016. A comprehensive generic-level phylogeny of the sunflower family:implications for the systematics of Chinese Asteraceae. J. Syst. Evol, 54: 416-437. DOI:10.1111/jse.12216 |
Freire S.E., 2017. Revision of the asian genus Pertya (Asteraceae, Pertyoideae). Syst. Bot. Monogr, 101: 1-90. |
Funk, V.A., Anderberg, A.A., Baldwin, B.G., Bayer, R.G., Bonifacino, J.M., Breitwieser, I., et al., 2009. Compositae metatrees: the next generation. In: Funk, V.A. (Ed.), Systematics, Evolution, and Biogeography of Compositae. IAPT, Vienna, pp. 747-777.
|
Gao, T.G., Nicholas, H.D., 2011. Myripnois Bunge. In: Wu, Z.Y., Raven, P.H., Hong, D.Y.(Eds.), Flora of China, vol. 21, pp. 31-32.
|
Girard S.L., Gauthier J., Noreau A., Xiong L., Zhou S., Jouan L., et al, 2011. Increased exonic de novo mutation rate in individuals with schizophrenia. Nat. Genet, 43: 860-863. DOI:10.1038/ng.886 |
George B., Bhatt B.S., Awasthi M., George B., Singh A.K., 2015. Comparative analysis of microsatellites in chloroplast genomes of lower and higher plants. Curr. Genet, 61: 665-677. DOI:10.1007/s00294-015-0495-9 |
Goulding S.E., Olmstead R.G., Morden C.W., Wolfe K.H., 1996. Ebb and flow of the chloroplast inverted repeat. Mol. Gen. Genet, 252: 195-206. DOI:10.1007/BF02173220 |
Govindaraj M., Vetriventhan M., Srinivasan M., 2015. Importance of the genetic diversity assessment in crop plants and its recent advances:an overview of its analytical perspectives. Genet. Res. Int, 43: 1487. DOI:10.1155/2015/431487 |
Huang L., Wang Z., Wang T., Su Y.J., 2016. The complete chloroplast genome sequence of Mikania micrantha (Asteraceae), a noxious invasive weed to South China. Mitochondrial DNA Part A, 1: 603-604. DOI:10.1080/23802359.2016.1209090 |
Hupfer H., Swiatek M., Hornung S., Herrmann R.G., Maier R.M., Chiu W.L., et al, 2000. Complete nucleotide sequence of the Oenothera elata plastid chromosome, representing plastome I of the five distinguishable Euoenothera plastomes. Mol. Genet. Genom, 263: 581-585. DOI:10.1007/PL00008686 |
Jansen R.K., Cai Z., Raubeson L.A., Daniell H., Leebens-Mack J., Müller K.F., et al, 2007. Analysis of 81 genes from 64 plastid genomes resolves relationships in angiosperms and identifies genome-scale evolutionary patterns. Proc. Natl. Acad. Sci, 104: 19369-19374. DOI:10.1073/pnas.0709121104 |
Jansen R.K., Palmer J.D., 1987. A chloroplast DNA inversion marks an ancient evolutionary split in the sunflower family (Asteraceae). Proc. Natl. Acad. Sci, 84: 581-582. DOI:10.1073/pnas.84.16.5818 |
Johansson J.T., 1999. There large inversions in the chloroplast genomes and one loss of the chloroplast gene rps16 suggest an early evolutionary split in the genus Adonis (Ranunculaceae). Plant Syst. Evol, 218: 133-143. DOI:10.1007/BF01087041 |
Katoh K., Toh H., 2010. Parallelization of the MAFFT multiple sequence alignment program. Bioinformatics, 26: 1899-1900. DOI:10.1093/bioinformatics/btq224 |
Kim H.G., Choi K.S., Jansen R.K., 2005. Two chloroplast DNA inversions originated simultaneously during the early evolution of the sunflower family (Asteraceae). Mol. Biol. Evol, 22: 1-10. DOI:10.1093/molbev/msi174 |
Kim K.A., Cheon K.S., Jang S.K., Yoo K.O., 2016. Complete chloroplast genome sequence of Adenophora remotiflora (Campanulaceae), Mitochondr. DNA Part A, 27: 2963-2964. DOI:10.3109/19401736.2015.1060461 |
Kurtz S., Choudhuri J.V., Ohlebusch E., Schleiermacher C., Stoye J., Giegerich R., 2001. REPuter:the manifold applications of repeat analysis on a genomic scale. Nucleic Acids Res, 29: 4633-4642. DOI:10.1093/nar/29.22.4633 |
Lanfear R., Calcott B., Ho Y.W., Guindon S., 2012. PartitionFinder:combined selection of partitioning schemes and substitution models for phylogenetic analyses. Mol. Biol. Evol, 29: 1695-1701. DOI:10.1093/molbev/mss020 |
Lee H.L., Jansen R.K., Chumley T.W., Kim K.J., 2007. Gene relocations within chloroplast genomes of Jasminum and Menodora (Oleaceae) are due to multiple, overlapping inversions. Mol. Biol. Evol, 24: 1161-1180. DOI:10.1093/molbev/msm036 |
Li H.T., Yi T.S., Gao L.M., Ma P.F., Zhang T., Yang J.B., et al, 2019. Origin of angiosperms and the puzzle of the Jurassic gap. Nat. Plants, 5: 461-470. DOI:10.1038/s41477-019-0421-0 |
Lin N., Moore M.J., Deng T., Sun H., Yang L.S., Sun Y.X., et al, 2018. Complete plastome sequencing from Toona (Meliaceae) and phylogenomic analyses within Sapindales. Appl. Plant Sci, 6: e1040. DOI:10.1002/aps3.1040 |
Lowe T.M., Eddy S.R., 1997. tRNAscan-SE:a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res, 25: 955-964. DOI:10.1093/nar/25.5.0955 |
Mandel J.R., Bayer M.S., Dikow R.B., Gao T.G., Jones K.E., Norbert S.K., et al, 2017. The Compositae tree of life in the age of phylogenomics. J. Syst. Evol, 55: 405-410. DOI:10.1111/jse.12265 |
Nie Z.L., Funk V.A., Meng Y., Deng T., Sun H., Wen J., 2015. Recent assembly of the global herbaceous flora:evidence from the paper daisies (Asteraceae:Gnaphalieae). New Phytol, 209: 1795-1806. DOI:10.1111/nph.13740 |
Oberprieler C., Himmelreich S., Vogt R., 2007. New subtribal classification of the tribe Anthemideae (Compositae). Willdenowia, 37: 89-114. DOI:10.3372/wi.37.37104 |
Panero J.L., Crozier B.S., 2016. Macroevolutionary dynamics in the early diversification of Asteraceae. Mol. Phylogenetics Evol, 99: 116-132. DOI:10.1016/j.ympev.2016.03.007 |
Panero J.L., Funk V.A., 2002. Toward a phylogenetic subfamily classification for the Compositae (Asteraceae). Proc. Biol. Soc, 115: 760-773. |
Panero J.L., Freire S.E., Espinar L.A., Crozier B.S., Barboza G.E., Cantero J.J., 2014. Resolution of deep nodes yields an improved backbone phylogeny and a new basal lineage to study early evolution of Asteraceae. Mol. Phylogenetics Evol, 80: 43-53. DOI:10.1016/j.ympev.2014.07.012 |
Panero J.L., Funk V.A., 2008. The value of sampling anomalous taxa in phylogenetic studies:major clades of the Asteraceae revealed. Mol. Phylogenetics Evol, 47: 757-782. DOI:10.1016/j.ympev.2008.02.011 |
Patel R.K., Jain M., 2012. NGS QC Toolkit:a toolkit for quality control of next generation sequencing data. PLoS One, 7: e30619. DOI:10.1371/journal.pone.0030619 |
Posada D., Crandall K.A., 1998. Modeltest:testing the model of DNA substitution. Bioinformatics, 14: 817-818. DOI:10.1093/bioinformatics/14.9.817 |
R Core Team, 2013. R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing, Vienna, Austria. http://www.R-project.org/.
|
Raubeson L.A., Peery R., Chumley T.W., Dziubek C., Fourcade N.M., Boore J.L., et al, 2007. Comparative chloroplast genomics:analyses including new sequences from the angiosperms Nuphar advena and Ranunculus macranthus. BMC Genomics, 8: 174-201. DOI:10.1186/1471-2164-8-174 |
Ronquist F., Teslenko M., van der Mark P., Ayres D.L., Darling A., 2012. MrBayes 3. 2:efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol, 61: 539-542. DOI:10.1093/sysbio/sys019 |
Rozas J., Sanchez-DelBarrio P., Messeguer X., Rozas R., 2003. DnaSP, DNA polymorphism analyses by the coalescent and other methods. Bioinformatics, 19: 2496-2497. DOI:10.1079/9780851994758.0139 |
Ruhfel B.R., Gitzendanner M.A., Soltis P.S., Soltis D.E., Burleigh J.G., 2014. From algae to angiosperms-inferring the phylogeny of green plants (Viridiplantae) from 360 plastid genomes. BMC Evol. Biol, 14: 23. DOI:10.1186/1471-2148-14-23 |
Shi C., Hu N., Huang H., Gao J., Zhao Y.J., Gao L.Z., 2012. An improved chloroplast DNA extraction procedure for whole plastid genome sequencing. PLoS One, 7: e31468. DOI:10.1371/journal.pone.0031468 |
Shinozaki K., Ohme M., Tanaka M., Wakasugi T., Hayshida N., Matsubayashi T., et al, 1986. The complete nucleotide sequence of the tobacco chloroplast genome:its gene organization and expression. EMBO J, 5: 2043-2049. DOI:10.1002/j.1460-2075.1986.tb04464.x |
Smith D.R., 2017. Does cell size impact chloroplast genome size? Front. Plant Sci, 8: 2116. DOI:10.3389/fpls.2017.02116 |
Sun Y.X., Moore M.J., Landis J.B., Lin N., Chen L., Deng T., et al, 2018. Plastome phylogenomics of the early-diverging eudicot family Berberidaceae. Mol. Phylogenetics Evol, 128: 203-211. DOI:10.1016/j.ympev.2015.12.006 |
Sun Y.X., Moore M.J., Lin N., Adelalu K.F., Meng A.P., Jian S.G., et al, 2017. Complete plastome sequencing of both living species of Circaeasteraceae (Ranunculales) reveals unusual rearrangements and the loss of the ndh gene family. BMC Genomics, 18: 592. DOI:10.1186/s12864-017-3956-3 |
Thiel T., Michalek W., Varshney R., Graner A., 2003. Exploiting EST databases for the development and characterization of gene-derived SSR-markers in barley(Hordeum vulgare L.). Theor. Appl. Genet, 106: 411-422. DOI:10.1007/s00122-002-1031-0 |
Vallès J., Torrell M., Garnatje T., Garcia-Jacas N., Vilatersana R., Susanna A., 2003. The genus Artemisia and its allies:phylogeny of the subtribe Artemisiinae(Asteraceae, Anthemideae) based on nucleotide sequences of nuclear ribosomal DNA internal transcribed spacers (ITS). Plant Biol, 5: 274-284. DOI:10.1055/s-2003-40790 |
Vargas O.M., Ortiz E.M., Simpson B.B., 2017. Conflicting phylogenomic signals reveal a pattern of reticulate evolution in a recent high-Andean diversification(Asteraceae:Astereae:Diplostephium). New Phytol, 214: 1736-1750. DOI:10.1111/nph.14530 |
Wang X.Y., Zhou Z.S., Liu G., Qian Z.Q., 2017. Characterization of the complete chloroplast genome of the invasive weed Galinsoga quadriradiata (Asterales:Asteraceae). Conserv. Genet. Res, 10: 89-92. DOI:10.1007/s12686-017-0771-8 |
Wei Z., Zhu X.S., Van den Berg R.G., Bakker F.T., Schranz M.E., 2015. Phylogenetic relationships within Lactuca L. (Asteraceae), including African species, based on chloroplast DNA sequence comparisons. Genet. Resour. Crop Evol, 64: 55-71. DOI:10.1007/s10722-015-0332-5 |
Wheeler G.L., Dorman H.E., Buchanan A., Challagundla L., Wallace L.E., 2014. A review of the prevalence, utility, and caveats of using chloroplast simple sequence repeats for studies of plant biology. Appl. Plant Sci, 2: 1400059. DOI:10.3732/apps.1400059 |
Wyman S.K., Jansen R.K., Boore J.L., 2004. Automatic annotation of organellar genomes with DOGMA. Bioinformatics, 20: 3252-3255. DOI:10.1093/bioinformatics/bth352 |
Xie Q.Z., Zhang X.H., 2012. Myripnois dioica:a beautiful woody dwarf plant material. J. Chin. Urban For, 10: 56-57. DOI:10.3969/j.issn.1672-4925.2012.05.021 |
Xue J., Wang S., Zhou S.L., 2012. Polymorphic chloroplast microsatellite loci in Nelumbo (Nelumbonaceae). Am. J. Bot, 99: e240-e244. DOI:10.3732/ajb.100547 |
Zhang N., Erickson D.L., Ramachandran P., Ottesen A.R., Timme R.E., Funk V.A., Luo Y., Handy S.M., 2017. An analysis of Echinacea chloroplast genomes:implications for future botanical identification. Sci. Rep, 7: 216. DOI:10.1038/s41598-017-00321-6 |
Zhang Y., Li L., Yan T.L., Liu Q., 2014. Complete chloroplast genome sequences of Praxelis (Eupatorium catarium Veldkamp), an important invasive species. Gene, 549: 58-69. DOI:10.1016/j.gene.2014.07.041 |



