b. University of Chinese Academy of Sciences, Beijing 100049, China;
c. Gardening and Horticulture Department, Xishuangbanna Tropical Botanical Garden, Chinese Academy of Sciences, Mengla, 666303, China;
d. Laboratory of Ecology and Evolutionary Biology, Yunnan University, Kunming, 650091, China
Flowers, the reproductive organs of angiosperms, display astonishing diversity in form. Understanding the variation of flower diversity has been an important research theme in biology since Darwin (Barrett, 2010). Some floral traits, such as floral shape and floral color, are more variable than others and tightly related with pollinator preference, possibly resulting in reproductive isolation and speciation (Hodges and Arnold, 1995; Classen-Bockhoff et al., 2003; Sheehan et al., 2016; Brock et al., 2016; Moyers et al., 2017).
Nectar spurs, the tubular outgrowths of petals or sepals, contain nectar rewards for pollinators. Spurs are thought to represent a key innovation in numerous angiosperm taxa because spur length variation allows pollinators to utilize resources in novel ways, leading to different selective pressures on spurs and allowing taxa to diversify (von Hagen and Kadereit, 2003; Young, 2008; Sletvold et al., 2010; Calcagno et al., 2017). For example, in Aquilegia plants, which have great interspecific variation in spur length, long spurs are currently under selection to match the length of pollinator mouthparts (Whittall and Hodges, 2007). Variations in nectar spur length strongly promote species diversity in Aquilegia and many other taxa of flowering plants (Hodges and Arnold, 1995; Fior et al., 2013). Thus, it would be especially interesting to understand the mechanisms responsible for spur formation.
|
Compared with our understanding of spur length and specialized pollinator interactions (Hodges, 1997; Whittall and Hodges, 2007), we know much less about the mechanisms involved in spur length and current advances have been mainly based on interspecific comparisons of cell morphology. Early anatomical evidence suggested that nectar spurs are derived from 'meristematic' bulges at the base of the petal (Erbar et al., 1999; Tucker and Hodges, 2005), and recent studies suggest that spur length is driven by cell divisions combined with anisotropic cell expansion (Puzey et al., 2012; Yant et al., 2015). Specifically, changes in anisotropic cell expansion rather than cell number have been shown to explain spur length diversity in Pelargonium, Gilia, Saltugilia and Centranthus (Tsai et al., 2018; Landis et al., 2016; Mack and Davis, 2015); a similar pattern may explain spur length diversity in the four Aquilegia species (Puzey et al., 2012). However, in other genera with spur length variation, such as Linaria, cell number has been found to be much higher in long-spurred species than in short-spurred species; furthermore, differences in cell length or cell anisotropy have been unable to explain these differences (Cullen et al., 2018). Clearly, the cellular and molecular mechanisms that regulate spur-length variation remain unresolved. Moreover, studies on intra-specific variation of spur length are rare, although floral traits of one species generally exhibit noticeable differences in different populations (Anderson et al., 2016; Yang et al., 2018). Thus, investigating intraspecific spur diversity may provide key insights into the underlying mechanisms of spur formation and variation.
Aquilegia rockii Munz is a small perennial herb that is native to southwest China. The spur-length of the species is normally distributed and ranges from 16 to 20 mm in most populations. However, during field survey we found an unusually variable population that exhibited a clinal variation in spur-length. Therefore, in the present research, we asked two questions. First, we asked whether changes in morphogenesis, e.g., cell number and/or size, are responsible for spur length in A. rockii. To answer this question, we measured the cell number and cell anisotropy of longand short-spurred flowers. Next, we asked whether genes associated with cell number or cell size differentially expressed between short- and long-spurred flowers. To answer this question, we examined the transcriptomic differences of key genes in both longand short-spurred flowers.
2. Material and methods 2.1. Plant materials and sample collectionA. rockii is a perennial herb, inhabiting the understory of mixed forests with a wide distribution in southwestern China. Each plant has a few ramets that develop into a cymose inflorescence with 1e15 pendulous purpose to blue flowers. In addition, there are 5 petals on each flower with a spur under each petal. In the study population on Hongshan mountain, Shangri-la, Yunnan, China (28°15'N, 100°5'E, 3600 asl.), spurs show great variation in length, bumblebees were principal pollinators in the population (Zhang ZQ, unpublished data). All samples were collected from this wild population under natural conditions, which does not require local and national permission (specimens were identified by Yan Luo and deposited at the Laboratory of Ecology and Evolutionary Biology, Yunnan University, China).
In the field, we randomly selected 100 individuals and measured spurs-length of four opened flowers from each individual. We found spur-length was multimodal with two peaks corresponding to long- and short-tubed phenotypes (L, 14.90 ± 0.08 mm and S, 3.10 ± 0.07 mm, respectively; mean ± SE) (Zhang ZQ, unpublished data). Spurs on three flowers at Stage 12 (Yant et al., 2015; Table S2) of each phenotype were collected and fixed with 2% glutaraldehyde in 0.2 M phosphate buffer for observation under the microscope. Additionally, from both long- and short-spurred plants, we collected newly opened flowers on three individuals, and immediately put them separately in liquid nitrogen for RNA extraction and sequencing.
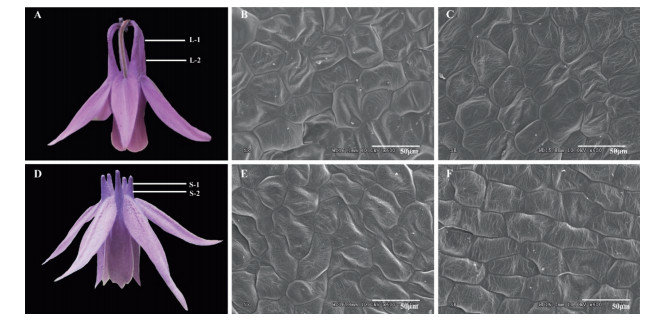
2.2. Scanning electron microscopyIn the laboratory, the spurs fixed in phosphate buffer were dehydrated in a graded alcohol series (from 30% to 100% for 20 min each) and finally soaked in tertiary butanol at -20℃. Samples were prepared for microscopy by a vacuum drying process and then incised at a width of 0.5 mm from one third and two thirds of whole spur length from the tip of the spur (Fig. 1A, D). After being coated with gold, the incised parts from the spur were scanned using a scanning electron microscope (SEM, 10 kV voltage, HITACHI S-3000N, Japan). For each section along spur at spur-length phenotype, two to four biological replicates were imaged at 600 × magnification, with a total of 330 distinct cell measurements. Cell density and cell anisotropy (cell length/cell width ratio) were quantified by ImageJ software v.1.48 (Collins, 2007). Specifically, cells per unit area (208 μm × 156 μm) were used to measure cell density; for these observations, at least two flowers were chosen, and a maximal length and width of five homogeneous cells were randomly selected for cell anisotropy measurement in each picture. To determine the differences in cell density and cell anisotropy between both the top and bottom of spurs and short- and longspurred flowers, we analyzed variance with SPSS software (IBM statistic 20), using position on spur and spur length as fixed factors.
|
| Fig. 1 Long- (A) and short-spurred flower (D) of Aquilegia rockii. Samples were collected by dissecting two parts of each flower for scanning electron microscopy: one third (1) and two thirds (2) of whole petal spur length, from the top to bottom position. Spur epidermis cells in L-1, top of long spur (B); L-2, bottom of long spur (C); S-1, top of short spur (E); S-2, bottom of short spur (F). |
RNA was extracted from triplicate pools of long- and shortspurred flowers, respectively. A total of six RNA samples (AL1, AL2, AL3, AS1, AS2 and AS3) were isolated using Eastep® super total RNA extraction kit according to manufacturer's protocols (Promega, Shanghai, China). RNA quality and quantity were estimated with 1% agarose gels and Agilent 2100 Bioanalyzer (Agilent Technologies, Santa Clara, USA). cDNA libraries were built with qualified RNA following Illumina's recommendations. RNA sequencing was conducted on an Illumina HiSeq2500 sequencer at Novogene Co, China.
2.4. De novo transcriptome assembly and clusteringFor all samples, raw reads were filtered to discard adaptor sequences and low-quality region reads. After trimming, these preprocessed reads were used as clean data sets which were provided in the NCBI SRA database (PRJNA506139). All clean reads were pooled and de novo transcriptome assembly was applied by Trinity software v.2.0.6 with default parameters (Grabherr et al., 2011). Clean reads were aligned with Bowtie 2 v.2.3.0 (Langmead and Salzberg, 2012) against the assembly sequence, allowing multimapping for each read. To obtain non-redundant reads and genelevel counts from each sample, Corset v.1.05 (Davidson and Oshlack, 2014) was utilized to hierarchically cluster the transcripts. Unigenes (representative transcripts) were generated based on the scale of shared reads.
2.5. Differential gene expression analysis between long- and shortspurred flowersDifferential expression analysis was performed in R using edgeR v.3.2.4 (Robinson et al., 2010). To minimize variance, genes less than 1 read per million (CPM) in at least 3 samples were excluded. Remaining genes were utilized to normalize and estimate the dispersion. The fold change value (log2FC), CPM value (log2CPM), P value and false discovery rate (FDR) for each transcript were measured. Heatmaps were created using CPM values. Differentially expressed genes were identified with the cutoff of FDR < 0.05 and |log2FC| > 2. To understand the major biological functions of differentially expressed genes, Gene Ontology (GO) annotations, enzyme annotations, and protein annotations were carried out by Blast2GO v.4.1 (Conesa et al., 2005) and Interproscan perl-based v.4.8 (Zdobnov and Apweiler, 2001) after BLASTX searches of the nr database (e value < 1e-5). KAAS (KEGG Automatic Annotation Server) was utilized to characterize the associated pathways of target genes (Moriya et al., 2007).
2.6. Quantitative real-time PCROnce RNA samples were sequenced, cDNA was synthesized with the GoScriptTM Reverse Transcription System Kit (Promega, USA) according to the manufacturer's instructions. Relative expression levels of genes related to cell division and anisotropic expansion were validated by quantitative real-time PCR (qRT-PCR), which was conducted with BrightGreen 2 × qPCR MasterMix-ROX kit (Applied Biological Materials Inc., Canada) and StepOneTM real-time PCR system (Applied Biosystems, USA). Primers were designed using ncbi primer-blast (Supplementary Table S1). The 20 μL PCR mixtures were composed of 0.6 μL forward and reverse primers at 10 μM, 10 μL BrightGreen 2 × qPCR MasterMix, and 2 μg of cDNA. All amplifications were pre-degenerated at 50℃ for 2 min, melted at 95℃ for 10 min, followed by 40 cycles of 95℃ for 15 s and 60℃ for 1 min. Reactions were run in three independent biological replicates for each gene. Relative gene abundance was calculated using the 2-△△CT method, with AqIPP2 as the internal control (Sharma et al., 2011). To evaluate differences in genes expression between long- and short-spurred flowers, we performed one-way ANOVA tests with SPSS software (IBM statistic 20).
3. Results 3.1. Cell morphology of long and short nectar spursSpur length of long- and short-spurred plants were significantly different; however, cellular morphology in long and short spurs were similar (Fig. 1). Cells at the top of both long and short spurs had irregular polygonal shapes that were not arranged uniformly (see Fig. 1, L-1 and S-1, one third of whole petal spur length). In contrast, near the bottom of both long and short spurs, cells were more regularly shaped and organized in neat rows (See Fig. 1, L-2 and S-2, two thirds of whole petal spur length).
Cell density and cell anisotropy did not differ significantly between long and short spurs (Fig. 2A). Specifically, cell density was similar at both the top and bottom of spurs in both long- and shortspurred plants [mean ± SE, 30.00 ± 0.00 vs 31.75 ± 1.65 in the bottom, and 32.50 ± 0.50 vs 29.00 ± 1.00 in the top position, F1, 7 = 0.08, P = 0.785]; furthermore, when we compared the cell density at the tops of long and short spurs, and at the bottoms of long and short spurs, we found no significant differences (Fig. 2A). Cell anisotropy did not differ significantly between long and short spurs (Fig. 2B). Specifically, there was no significant difference in cell anisotropy between the top and bottom of the spur in either long- or short-spurred plants; in addition, we found no differences in cell anisotropy when we compared the spur tops of long and short spurs, or the spur bottoms of each spur type [mean ± SE, 1.15 ± 0.05 vs 1.32 ± 0.06 in the bottom, and 1.34 ± 0.11 vs 1.14 ± 0.05 in the top position, F1, 17 = 0.023, P = 0.882].

|
| Fig. 2 Cell numbers (A) and anisotropy (B) of long- (grey) and short-spurred (white) flowers under unit size. L-1, top of long spur; L-2, bottom of long spur; S-1, top of short spur; S- 2, bottom of short spur. Sample sizes are 65, 132, 70, and 63 in the S-1, S-2, L-1, and L-2, respectively. |
Using next-generation sequencing, we generated a total of 65, 727, 602 and 65, 029, 977 short reads for long- (AL) and shortspurred (AS) flowers, respectively. Of these reads, most had quality scores above 98.5%, indicating that sequencing data were accurate enough for further analysis. The raw reads were separately trimmed, retaining 64, 713, 424 and 62, 612, 564 reads for AL and AS samples, covering 98.46% and 96.28%, respectively. After that, the clean reads were pooled and assembled using the Trinity software to obtain a uniform reference sequence. Altogether 573, 096 transcripts were obtained, with the N50 value as 601 (Table 1). Corset program was used to identify redundant and similar transcripts. Correspondingly, high-quality reference sequences with a total of 58, 973 clusters were procured. The non-redundant reads were defined as unigenes and subjected to further analysis.
| Item | Raw reads | Clean reads | Effective Rate (%) | GC Content(%) |
| AL1 | 24, 594, 136 | 24, 393, 124(99.18%) | 99.17 | 41.64 |
| AL2 | 20, 333, 334 | 20, 033, 334(98.52%) | 98.94 | 41.36 |
| AL3 | 20, 800, 132 | 20, 286, 966(97.53%) | 99.53 | 40.41 |
| AS1 | 21, 823, 851 | 21, 599, 896(98.97%) | 98.96 | 41.41 |
| AS2 | 20, 965, 738 | 20, 725, 702(98.86%) | 98.86 | 41.06 |
| AS3 | 22, 240, 388 | 21, 956, 286(98.72%) | 98.72 | 41.33 |
| Altogether | 130, 757, 57 | 128, 995, 308(98.65%) | ||
| N50 | 601 | |||
| Number of transcripts after assembly | 573, 096 | |||
| Number of unigenes after clustering | 58, 973 | |||
| AS1-3, cDNA from three independent biological samples of short-spurred flowers; AL1-3, cDNA from three independent biological samples of long-spurred flowers. | ||||
To identify differentially expressed genes in A. rockii, cluster analyses were performed using CPM values. In total, 672 genes displayed significantly differential expression (FDR < 0.05) in AL vs AS comparison. Of these, most transcripts were upregulated in AS samples, while a smaller number of transcripts (199 genes) were down-regulated (Supplementary Figure S1). The high expression genes in AL samples may devote to the long spur phenotype.
To investigate the function of differentially expressed genes, we annotated these genes using GO, KEGG, and Interproscan. According to blast results, differentially expressed genes from short- and long-spurred flowers were most frequently matched with genes from Nelumbo nucifera (Supplementary Figure S2). Furthermore, the most significantly differentially expressed genes were genes homologous to F-box, Cyclin-dependent kinase, and LST8 (log2FC > 4) (Supplementary Table S2). In the GO annotation, differentially expressed genes were categorized into 464 functional categories, including molecular function (MF, 207, 44.61%), biological process (BP, 191, 41.16%), and cellular components (CC, 66, 14.23%) (Supplementary Table S3). In general, the majority of GO annotations for both up-regulated and down-regulated genes fell into the category of MF, followed by BP, and CC. Specifically, oxidoreductase activity, oxidation-reduction process, and membrane represented the dominant GO terms for differentially expressed genes in the MF, BP and CC subgroups, individually (Supplementary Figure S3).
The top 12 KEGG pathways were shown in Supplementary Figure S4. Among these pathways, a majority of genes appeared to play a role in metabolic pathways (27.27%), followed by biosynthesis of secondary metabolites (13.64%). Differentially expressed genes were also annotated with Interproscan (Supplementary Table S4). Among all the differentially expressed genes, 141 were annotated as enzymes (Supplementary Table S5). The Interproscan and enzyme results indicate that these differentially expressed genes may have many functions, which has helped us screen for key genes involved in intra-specific spur length variation.
3.4. Differentially expressed genes related to cell numberScanning electron microscopy revealed that there were no differences in either cell density or cell anisotropy between short- and long-spurred flowers, suggesting that in A. rockii changes in cell number may explain variations in spur length. Therefore, we focused our attention on gene expression that might be linked to changes in cell number. Specifically, the gene with the highest level of differential expression in long-spurred flowers encodes an F-box gene with an NHL repeat domain and WD40 domain (Supplementary Figure S5). Furthermore, several genes associated with cell division processes, e.g., a cyclin-dependent kinase gene (CDKB2-2) and LST8 gene, were down-regulated in short-spurred flowers (Fig. 3; Supplementary Table S6). We also checked the expression patterns of genes involved in hormone transduction. Our analyses did not identify differentially expressed genes that are involved in cell division-linked hormone signaling transduction pathways, such as the auxin synthetic, brassinosteroid, or cytokinin pathways. Thus, cell number changes in spur length variation appear to occur through a hormone-independent pathway.
|
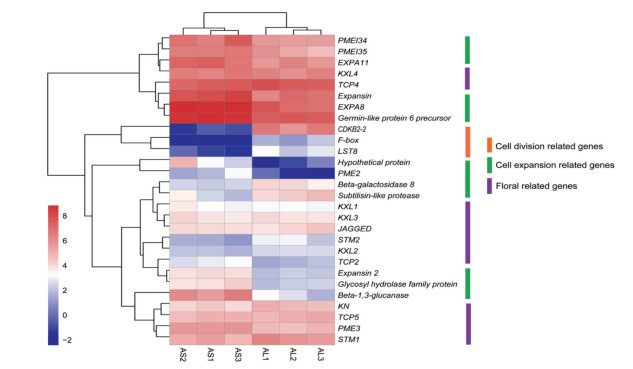
| Fig. 3 Gene expression heatmap of candidate differentially expressed genes associated with cell division, cell expansion, and flowers in Aquilegia rockii. Red color boxes represent high gene expression, white and blue boxes represent relatively low expression. The orange line represents cell division-related genes; green and purple lines represent genes associated with cell expansion and flowers. AS1-3, cDNA from three independent biological samples of short-spurred flowers; AL1-3, cDNA from three independent biological samples of long-spurred flowers. |
We speculated that the special structure of spurs requires differential expression of genes in the cell wall and plant-type cell wall GO subcategories. Our analyses revealed 14 cell wall genes that were differentially expressed, including the alpha-expansin family protein (EXPA), pectinesterase family protein (PME), pectinesterase inhibitor family protein (PMEI), and other proteins (Fig. 3; Supplementary Table S7). Most genes related with anisotropic cell expansion were slightly upregulated in AS samples, indicating that these genes play a role in anisotropic cell expansion and short spur formation.
To further explore the expression patterns of well-studied floralspecific genes in A. rockii, the homologues of KNOX, TCP4 and JAGGED were identified through blast searches against published data of A. coerulea. Also, the homologues of TCP2 and TCP5 were identified by blast searches against Arabidopsis thaliana. Altogether, 7 KNOX, 3 TCP, and a JAGGED homologues were identified. However, these genes were not differentially expressed between long- and short-spurred flowers (Fig. 3; Supplementary Table S8).
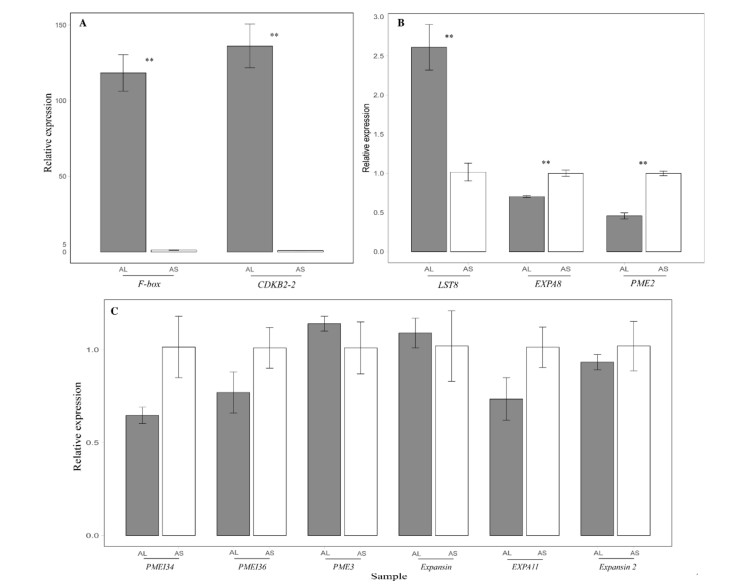
3.6. Quantitative real-time PCR of candidate genesBased on our transcriptome analysis, we selected 12 transcripts related to cell division and anisotropic cell expansion to screen using qRT-PCR. Excluding Pectinesterase 3 (PME3) and Expansin, all transcripts showed similar expression levels as the RNA-seq results (Fig. 4; Supplementary Table S6; Supplementary Table S7). The expression levels of three genes related to cell division (i.e., F-box, CDKB2-2, and LST8) increased greatly in AL sample. In contrast, limited cell expansion genes (i.e., Expansin-A8 like precursor (EXPA8), Pectinesterase 2 (PME2)) showed apparent increasing expression tendency in AS samples. However, the expression of most expansion genes, such as Pectinesterase inhibitor 34 (PMEI34), Pectinesterase inhibitor 36 (PMEI36), alpha-expansin 11 precursor (EXPA11), showed no notable change in short-spurred flowers.
|
| Fig. 4 Quantitative real-time PCR analysis between long- (grey) and short-spurred (white) flowers (means ± SE). Bars with double asterisks indicate significant difference at the 0.01 level between the long- and short-spurred flowers. AS1-3, cDNA from three independent biological samples of short-spurred flowers; AL1-3, cDNA from three independent biological samples of long-spurred flowers. |
The formation and development of nectar spur involves two coordinated processes: an increase in cell number through cell division at early stage and subsequent cell expansion through an increase in cell anisotropy. Accordingly, changes in cell number and cell anisotropy may account for spur length diversity nonexclusively. Presently, studies of spur length variation have mainly focused on inter-species variation. This cell expansion through an increase in cell anisotropy is widely responsible for tube-length or spur-length divergences at the species level and above. For example, in Aquilegia, cell number accounts for less than 1% of spurlength variation, whereas changes in cell anisotropy determine the plasticity of inter-specific spur length (Tucker and Hodges, 2005). The change in cell anisotropy has also been found in Pelargonium, Gilia, Saltugilia, and Centranthus (Mack and Davis, 2015; Landis et al., 2016; Tsai et al., 2018). However, in the long-spurred Linaria becerrae, cell number is much higher than that in short-spurred L. clementei, but cell length and cell anisotropy have not been not found to change much (Cullen et al., 2018). Accordingly, both cell number and cell anisotropy may contribute to final spur length, depending on plant species. However, little is known about the mechanisms that underlie intra-specific variation of spur length. In contrast with most previous studies, our results show that cell number but not cell anisotropy, determines spur-length variation in A. rockii. This finding is supported by two lines of evidence. Firstly, we found that cell morphology was not significantly different between long- and short-spurred flowers at different positions of spurs (Fig. 1), and neither cell density nor the cell anisotropy provide support for spur length variation of A. rockii (Fig. 2). Therefore, spur length differences between long- and short-spurred flowers may mainly result from changes in cell number. Our results are in accordance with previous studies in A. coerulea that spur development is primarily driven by cell division (Yant et al., 2015).
Secondly, our RNA-seq results indicate that genes linked to cell division display differential expression between long- and shortspurred flowers. Our transcriptomic data and quantitative real-time PCR screen revealed significant down-regulation of genes associated with cell division (i.e., an F-box gene with an NHL repeat domain, CDKB2-2, and LST8) in short-spurred flowers (Figs. 3 and 4; Supplementary Table S6). Previous studies have indicated these down-regulated genes participate in cell number changes. For example, Wang et al. (2016) found that F-box domain genes play vital roles in controlling organ size by promoting the proliferation of meristemoid cells. In addition, proteins with NHL repeat domains are known to have a cell division functions, thus supporting the findings in rice that the NHL repeat domain plays a vital role in determining organ size (Chen et al., 2015). Also, our finding that CDKB2-2 and LST8 are down-regulated in short-spurred flowers suggests that these genes play a role in controlling spur length. This interpretation is in accordance with previous studies that CDKB2 is required for cell cycle regulation and meristem organization (Andersen et al., 2008), and LST8 is involved in adventitious root formation (Deng et al., 2017). The increased expression of cell division related genes in long-spurred flowers indicates that hormone-independent pathways may be crucial for increasing cell numbers and forming long spurs.
The genetic control of cell number in morphogenesis has been extensively studied. Previous research has suggested that genes involved in hormone-related pathways mainly contribute to the timing of cell proliferation. For instance, in A. coerulea, auxin- and brassinosteroid signaling-related genes (e.g., TCP4, CYP71A, ARF8, SAUR) show differential expression during the spur development process (Yant et al., 2015). Specifically, among these hormonerelated genes, TCP4 has been shown to restrain cell division, whereas CYP71A, ARF8, and SAUR have been shown to be highly expressed during the stages that correspond to spur development. However, in our study, hormone-related genes were not differentially expressed between long- and short-spurred flowers, indicating that cell division in spur formation may occur through a hormone-independent pathway. In addition to cell division, genes that modify anisotropic cell expansion and cell division duration may also influence organ size (Box et al., 2011; Li et al., 2016; Moyroud and Glover, 2017). However, in our study, most genes related to anisotropic cell expansion and cell division duration (e.g., genes of the Expansin, PME, and KNOX family) showed no differential expression between long- and short-spurred flowers (Fig. 4; Supplementary Table S8). Only two genes, encoding the proteins EXPA8 and PME2, showed increased expression in short-spurred samples. These limited cell expansional contribution can be explained for the organ compensation measurements in different sizes, such as leaf size coordination (Horiguchi et al., 2006). Nevertheless, we speculate that after cells reach a balance between division and expansion the genes involved in cell number changes play a more central role in spur formation than in cell expansion. Our finding that genes associated with cell division are expressed at significantly higher levels in long-spurred flowers implies that these genes contribute to large cell numbers during the formation of long spurs. In addition, our gene expression results are in general agreement with the SEM data that a change in cell number is a major factor in spur length variation of A. rockii.
5. ConclusionsThe ecological importance of spur diversity has been studied extensively in Aquilegia, but the mechanisms underlying the intraspecific variations of spur length still remain largely unclear. To the best of our knowledge, our study is the first attempt to identify both cellular processes and genes associated with intraspecific changes in spur length in A. rockii. Our results suggest that (1) the variation in intraspecific spur length in A. rockii is primarily driven by cell number, and that (2) the genetic pathways that regulate this process are not hormone-related but associated with cell division. These findings indicate that the cellular mechanisms that drive intraspecific spur variation differ from the those that drive interspecific spur length in Aquilegia. Although further studies in A. rockii are required to elucidate the spatiotemporal coordination of cell division and anisotropic cell expansion during spur development and should investigate the spatiotemporal expression patterns of newly identified candidate genes, our findings provide new insight into the mechanisms of spur diversification.
Conflicts of interestThe author declares no conflict of interest.
Author contributionsYD, YY, and ZQZ conceived and designed the experiments. ZlZ and YL collected the materials. ZlZ, YD, ZQZ performed the experiments. ZlZ and ZQZ interpreted the data. ZlZ, YD, ZQZ, and YL wrote the manuscript. All authors reviewed and approved the manuscript.
Data accessibilityThe transcriptome data used for this study were deposited at the National Center for Biotechnology Information Short Read Archive (http://www.ncbi.nlm.nih.gov/sra/) under the accession number PRJNA506139.
AcknowledgementsWe thank Dr. Wen Guo for field sampling. This work was financially supported by National Natural Science Foundation of China (31760104, 41461014, 31460040, and 31870183).
Appendix A. Supplementary dataSupplementary data to this article can be found online at https://doi.org/10.1016/j.pld.2019.06.001.
Andersen S.U., Buechel S., Zhao Z., et al, 2008. Requirement of B2-type cyclindependent kinases for meristem integrity in Arabidopsis thaliana. Plant Cell, 20: 88-100. DOI:10.1105/tpc.107.054676 |
Anderson B., Pauw A., Cole W.W., et al, 2016. Pollination, mating and reproductive fitness in a plant population with bimodal floral-tube length. J. Evol. Biol, 29: 1631-1642. DOI:10.1111/jeb.12899 |
Barrett S.C.H., 2010. Darwin's legacy: the forms, function and sexual diversity of flowers. Phil. Trans. Roy. Soc. Lond. B, 365: 351-368. DOI:10.1098/rstb.2009.0212 |
Box M.S., Dodsworth S., Rudall P.J., et al, 2011. Characterization of Linaria KNOX genes suggests a role in petal-spur development. Plant J, 68: 703-714. DOI:10.1111/j.1365-313X.2011.04721.x |
Brock M.T., Lucas L.K., Anderson N.A., et al, 2016. Genetic architecture, biochemical underpinnings and ecological impact of floral UV patterning. Mol. Ecol, 25: 1122-1140. DOI:10.1111/mec.13542 |
Calcagno V., Jarne P., Loreau M., et al, 2017. Diversity spurs diversification in ecological communities. Nat. Commun, 8: 15810-15818. DOI:10.1038/ncomms15810 |
Chen J., Gao H., Zheng X.M., et al, 2015. An evolutionarily conserved gene, FUWA, plays a role in determining panicle architecture, grain shape and grain weight in rice. Plant J, 83: 427-438. DOI:10.1111/tpj.12895 |
Classen-Bockhoff R., Wester P., Tweraser E., 2003. The staminal lever mechanism in Salvia L. (Lamiaceae) - a review. Plant Biol, 5: 33-41. DOI:10.1055/s-2003-37973 |
Collins T.J., 2007. ImageJ for microscopy. Biotechniques, 43: 25-30. DOI:10.2144/000112505 |
Conesa A., Gotz S., Garcia-Gomez J.M., et al, 2005. Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics, 21: 3674-3676. DOI:10.1093/bioinformatics/bti610 |
Cullen E., Fernandez-Mazuecos M., Glover B.J., 2018. Evolution of nectar spur length in a clade of Linaria reflects changes in cell division rather than in cell expansion. Ann. Bot, 122: 801-809. |
Davidson N.M., Oshlack A., 2014. Corset: enabling differential gene expression analysis for de novo assembled transcriptomes. Genome Biol, 15: 410-423. |
Deng K.X., Dong P., Wang W.J., et al, 2017. The TOR pathway is involved in adventitious root formation in Arabidopsis and Potato. Front. Plant Sci, 8: 784-801. DOI:10.3389/fpls.2017.00784 |
Erbar C., Kusma S., Leins P., 1999. Development and interpretation of nectary organs in Ranunculaceae. 194 :317-332.. New Flora, 194: 317-332. DOI:10.1016/S0367-2530(17)30920-9 |
Fior S., Li M., Oxelman B., et al, 2013. Spatiotemporal reconstruction of the Aquilegia rapid radiation through next-generation sequencing of rapidly evolving cpDNA regions. New Phytol, 198: 579-592. DOI:10.1111/nph.12163 |
Grabherr M.G., Haas B.J., Yassour M., et al, 2011. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol, 29: 644-652. DOI:10.1038/nbt.1883 |
Hodges S.A., 1997. Floral nectar spurs and diversification. Int. J. Plant Sci, 158: S81-S88. DOI:10.1086/297508 |
Hodges S.A., Arnold M.L., 1995. Spurring plant diversification: are floral nectar spurs a key innovation? Proc. R. Soc. B Biol. Sci, 262: 343-348. DOI:10.1098/rspb.1995.0215 |
Horiguchi G., Ferjani A., Fujikura U., et al, 2006. Coordination of cell proliferation and cell expansion in the control of leaf size in Arabidopsis thaliana. J. Plant Res, 119: 37-42. DOI:10.1007/s10265-005-0232-4 |
Landis J.B., O'Toole R.D., Ventura K.L., et al, 2016. The phenotypic and genetic underpinnings of flower size in Polemoniaceae. Front. Plant Sci, 6: 1144-1163. |
Langmead B., Salzberg S.L., 2012. Fast gapped-read alignment with Bowtie 2. Nat. Methods, 9: 357-359. DOI:10.1038/nmeth.1923 |
Li Y., Tu L.L., Pettolino F.A., et al, 2016. GbEXPATR, a species-specific expansin, enhances cotton fibre elongation through cell wall restructuring. Plant Biotechnol. J, 14: 951-963. DOI:10.1111/pbi.12450 |
Mack J.L.K., Davis A.R., 2015. The relationship between cell division and elongation during development of the nectar-yielding petal spur in Centranthus ruber(Valerianaceae). Ann. Bot, 115: 641-649. DOI:10.1093/aob/mcu261 |
Moriya Y., Itoh M., Okuda S., et al, 2007. KAAS: an automatic genome annotation and pathway reconstruction server. Nucleic Acids Res, 35: W182-W185. DOI:10.1093/nar/gkm321 |
Moyers B.T., Owens G.L., Baute G.J., et al, 2017. The genetic architecture of UV floral patterning in sunflower. Ann Bot, 120: 39-50. DOI:10.1093/aob/mcx038 |
Moyroud E., Glover B.J., 2017. The evolution of diverse floral morphologies. Curr. Biol, 27: R941-R951. DOI:10.1016/j.cub.2017.06.053 |
Puzey J.R., Gerbode S.J., Hodges S.A., et al, 2012. Evolution of spur-length diversity in Aquilegia petals is achieved solely through cell-shape anisotropy. Proc. R. Soc. B Biol. Sci, 279: 1640-1645. DOI:10.1098/rspb.2011.1873 |
Robinson M.D., McCarthy D.J., Smyth G.K., 2010. edgeR: a bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics, 26: 139-140. DOI:10.1093/bioinformatics/btp616 |
Sharma B., Guo C., Kong H., et al, 2011. Petal-specific subfunctionalization of an APETALA3 paralog in the Ranunculales and its implications for petal evolution. New Phytol, 191: 870-883. DOI:10.1111/j.1469-8137.2011.03744.x |
Sheehan H., Moser M., Klahre U., et al, 2016. MYB-FL controls gain and loss of floral UVabsorbance, a key trait affecting pollinator preference and reproductive isolation. Nat. Genet, 48: 159-166. DOI:10.1038/ng.3462 |
Sletvold N., Grindeland J.M., Agren J., 2010. Pollinator-mediated selection on floral display, spur length and flowering phenology in the deceptive orchid Dactylorhiza lapponica. New Phytol, 188: 385-392. DOI:10.1111/j.1469-8137.2010.03296.x |
Tsai T., Diggle P.K., Frye H.A., et al, 2018. Contrasting lengths of Pelargonium floral nectar tubes result from late differences in rate and duration of growth. Ann. Bot, 121: 549-560. DOI:10.1093/aob/mcx171 |
Tucker S.C., Hodges S.A., 2005. Floral ontogeny of Aquilegia, Semiaquilegia, and Enemion (Ranunculaceae). Int. J. Plant Sci, 166: 557-574. DOI:10.1086/429848 |
von Hagen K.B., Kadereit J.W., 2003. The diversification of Halenia (Gentianaceae):ecological opportunity versus key innovation. Evolution, 57: 2507-2518. DOI:10.1111/j.0014-3820.2003.tb01495.x |
Wang Z., Li N., Jiang S., et al, 2016. SCFSAP controls organ size by targeting PPD proteins for degradation in Arabidopsis thaliana. Nat. Commun, 7: 11192-11202. DOI:10.1038/ncomms11192 |
Whittall J.B., Hodges S.A., 2007. Pollinator shifts drive increasingly long nectar spurs in columbine flowers. Nature, 447: 706-709. DOI:10.1038/nature05857 |
Yang M.L., Wang L.L., Zhang G.P., et al, 2018. Equipped for migrations across high latitude regions? Reduced spur length and outcrossing rate in a biennial Halenia elliptica (Gentianaceae) with mixed mating system along a latitude gradient. Front. Genet, 9: 223-229. DOI:10.3389/fgene.2018.00223 |
Yant L., Collani S., Puzey J., et al, 2015. Molecular basis for three-dimensional elaboration of the Aquilegia petal spur. Proc. R. Soc. B Biol. Sci, 282: 20142778. DOI:10.1098/rspb.2014.2778 |
Young H.J., 2008. Selection on spur shape in Impatiens capensis. Oecologia, 156: 535-543. DOI:10.1007/s00442-008-1014-1 |
Zdobnov E.M., Apweiler R., 2001. InterProScan - an integration platform for the signature-recognition methods in InterPro. Bioinformatics, 17: 847-848. DOI:10.1093/bioinformatics/17.9.847 |



