b. Institute of Life Sciences, NALCO Nagar Road, NALCO Square, Chandrasekharpur, Bhubaneswar, 751023, India;
c. Centre for Rural Technology, Indian Institute of Technology Guwahati, Assam, 781 039, India;
d. Key Laboratory for Plant Diversity and Biogeography of East Asia, Kunming Institute of Botany, Chinese Academy of Sciences, Kunming 650201, China
Plants have been used since prehistoric times to treat serious ailments. Ayurveda, which is the traditional Hindu system of medicine, utilizes more than 5, 000 plant species (Adyanthaya et al., 2016). Kaempferia galanga(Zingiberaceae), which is used in 59 different Ayurvedic medicines(Sivarajan and Balachandran, 1994), has become extremely valuable; from 2014 to 2016, the price of this essential herb tripled in India (Preetha et al., 2016). However, the ever-increasing demand for traditional medicine has led to adulteration of Zingiberaceae in the international and national herbal market. For example, in Kerala, India Kaempferia rotunda(Zingiberaceae), the major ingredient of the popular Ayurvedic drug "Hallakam, " which is used to treat stomach aches, wounds, mental disorders, and insomnia, is commonly adulterated by Lagenandra toxicaria(Sereena et al., 2011). Consumers are often unaware of this adulteration and buy herbs that have a similar appearance to authentic Kaempferia species (Sasikumar et al., 2016).
Previous research has identified common adulterants of Ayurvedic drugs in India (Kumar and Ruba, 2018). During an ethno-medicinal survey in Northeast (NE) India, we found that Kaempferia species are adulterated by species belonging to the Marantaceae family (e.g Calathea bachemiana and Maranta leuconeura). An additional report also supports our field observation that species from Zingiberaceae (Curcuma angustifolia Roxb.) and Marantaceae (Maranta arundinacea Linn.) are used interchangeably in the Ayurvedic drug "Tugaksheeree" which aids digestion and metabolism (Rajashekhara and Sharma, 2010). When herbal healers misidentify Zingiberaceae plants, the therapeutic efficiency of Kaempferia-derived Ayurvedic drugs (e.g., Asaneladi Velichenna, Agasthyarasayanam, Eladi Velichenna, Gandha Thailam, Triphaladi Thailam, Rasnairandadi Kashayam) is markedly reduced. This taxonomic confusion is related to the rapid radiation in the order Zingiberales (Barrett et al., 2014), which consists of eight families (Marantaceae, Cannaceae, Zingiberaceae, Costaceae, Heliconiaceae, Strelitziaceae, Musaceae, and Lowiaceae) that share similar characteristics. Specifically, plants of these families are rhizomatous herbs, with midribs, ligules, involucral bracts, zygomorphic flowers, which have stamens and filaments with a staminoid structure (Carlsen et al., 2018). Furthermore, the flowering season of Kaempferia is short, and inflorescences of some species (e.g., K. rotunda) appear before leafy shoots and last only for 1–2 weeks. The absence of floral parts for most of the year and the dormancy throughout the non-rainy season make identifying Kaempferia more difficult. Because the preparation of authentic herbal medicine from Kaempferia requires accurate discrimination of adulterants from the pure Kaempferia complex, investigating effective methods of identifying Kaempferia and its adulterant species is crucial.
An accurate marker system that allows non-specialists to scrutinize the purity of the voucher is necessary. When the vouchers for herbal plants are processed into powder form, traditional organoleptic and elemental compositional analysis are often not sufficient to discriminate adulterants. Aside from using high performance chromatography to determine the presence of major compounds (Septyanti et al., 2016), molecular methods to authenticate Kaempferia-based herbal products remain scarce. One promising approach in detecting desired plants from the herbal formulation is DNA barcoding (Newmaster et al., 2013). Barcoding has effectively discriminated adulterants from industrial olive oil (Kumar et al., 2011), tea packets (Stoeckle et al., 2011), and turmeric(Parvathy et al., 2014). Plant barcodes proposed by the Consortium for the Barcode of Life's Plant Working Group include seven loci (rpoB, rpoC1, rbcL and matK, atpF-atpH, psbK-psbI and trnH-psbA) (CBOL Plant Working Group, 2009). However, it is unclear whether these barcodes individually or in combination are effective at discriminating individual families of plant species. For example, previous studies on Kaempferia phylogeny have attempted to use barcodes that consisted of single loci (petA-psbJ, trnH-psbA), two-locus concatenations combinations (Techaprasan et al., 2010). Efforts to detect adulteration in Kaempferia, however, have been scarce. Thus, we aimed to determine which of the DNA barcodes proposed by the CBOL's Plant Working Group are the most effective at discriminating Kaempferia from its adulterants.
In this study, we assessed the effectiveness of eight plastid loci (accD, rpoB, rpoC1, rbcL and matK, atpF-atpH, psbK-psbI and trnH-psbA) in discriminating adulterants from the Kaempferia species complex. We also propose the best barcode to authenticate Kaempferia species from their adulterants.
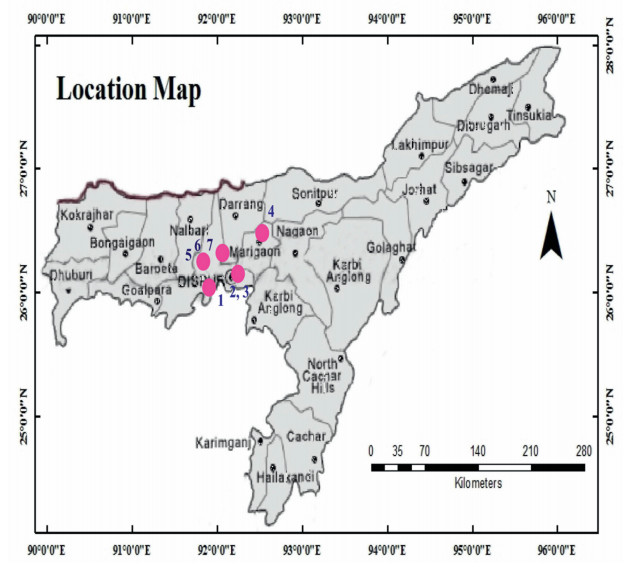
2. Materials and methods 2.1. Plant materialPlants belonging to Zingiberaceae and Marantaceae are the dominant food source of elephants in the forests of Assam. Taxonomically, plants of the Marantaceae family are poorly investigated in Asia; previous studies on Marantaceae reported 55 species throughout Asia (Suksathan et al., 2009). The species from the Marantaceae and Zingiberaceae families propagate clonally with rhizomes. To proliferate rapidly, the above ground biomass for species from both families has a very high growth rate. Furthermore, plants of the families Marantaceae and Zingiberaceae become reproductively active during the monsoon. When the floweringseason is over, the morphological similarity between the leaves of members of both families make identification at the species level difficult for herbal healers. This difficulty in identification, in turn, increases adulteration of Kaempferia by Marantaceae species with similar leaf morphology. To sort out this issue, the species from both families were collected during the monsoon season and identified by a renowned curator of Gauhati University, Dr. Gajen Chandra Sarma. The materials for the present study consisted of four species of Kaempferia and two species of family Marantaceae collected from Assam (Supplementary Table 1, Fig. 1). Two species (C. bachemiana E. Morren, M. leuconeura E. Morren 1, M. leuconeura E. Morren 2) that are commonly used as adulterator were also selected (Fig. 2). The species were maintained in the Departmental green house of IIT Guwahati, Assam.
| Primer name | Primer sequence | PCR conditions |
| rpoC1 | F – GGCAAAGAGGGAAGATTTCG R – CCATAAGCATATCTTGAGTTGG |
95 ℃-5 min; 30 cycles- 94 ℃-1 min, 57 ℃ -30 sec, 72 ℃ -45 sec; 72 ℃ -7 min |
| rpoB | F – ATGCAACGTCAAGCAGTTCC R – CCGTATGTGAAAAGAAGTATA |
95 ℃-5 min; 30 cycles- 94 ℃-1 min, 57 ℃ -30 sec, 72 ℃ -45 sec; 72-0C -7 min |
| matK | F – CGTACAGTACTTTTGTGTTTACGAG R – ACCCAGTCCATCTGGAAATCTTGGTTC |
95 ℃-4 min; 30 cycles- 94 ℃-30sec, 50 ℃ -1min, 72 ℃ -40 sec; 72 ℃ -7 min |
| rbcL | F – ATGTCACCACAAACAGAGACTAAAGC R – GTAAAATCAAGTCCACCRCG |
95 ℃-4 min; 30 cycles- 94 ℃-30 sec, 55 ℃ -1min, 72 ℃ -1 min; 72 ℃ -10 min |
| accD | F – CTATAGCAATTGGAGTTATGAATT R – CGGATCAATCAAAAGTTCGAT TC |
95 ℃-4 min; 30 cycles- 940C-30 sec, 50 ℃ -1 min, 72 ℃ -40 sec; 72 ℃ -7 min |
| atpF-atpH | F – ACTCGCACACACTCCCTTTCC R – GCTTTTATGGAAGCTTTAACAAT |
95 ℃-4 min; 30 cycles- 94 ℃-30 sec, 50 ℃ -1 min, 72 ℃ -40 sec; 72 ℃ -7 min |
| psbK-psbI | F – TTAGCCTTTGTTTGGCAAG R – AGAGTTTGAGAGTAAGCAT |
95 ℃-4 min; 30 cycles- 94 ℃-30 sec, 50 ℃ -1min, 72 ℃ -40 sec; 72 ℃ -7 min |
| trnH-psbA | F – GTTATGCATGAACGTAATGCTC R – CGCGCATGGTGGATTCACAATCC |
95 ℃-4 min; 30 cycles- 94 ℃-30 sec, 55 ℃ -1min, 72 ℃ -1 min; 72 ℃ -10 min |
|
| Fig. 1 Collection sites of different Kaempferia and their adulterant species used in the current study. Samples have been shown in the figures by their serial number given in Supplementary Table 1. 1. K. galanga, 2. K. angustifolia, 3. K. elegans, 4. K. rotunda, 5. C. bachemiana, 6. M. leuconeura 1, 7. M. leuconeura 2. |

|
| Fig. 2 Morphological variation in leaves (dorsal and ventral side) from Kaempferia complex and their adulterants. (a) K. galanga, (b) K. angustifolia (c) K. elegans, (d) K. rotunda, (e) C. bachemiana (f) M. leuconeura 1. |
Total genomic DNA was extracted from fresh tender leaves of the material using a DNeasy Plant mini kit (Qiagen Inc., Venlo, Netherlands). The quality and quantity of the extracted DNA was confirmed by running the extracted DNA on a 0.8% agarose gel, stained with ethidium bromide(0.5 μg μL−1). The universal primers of eight plastid loci used to discriminate the Kamepferia species from its adulterants were amplifed using the coding regions of ribulose-1, 5-bisphosphatecarboxylase/oxygenase large subunit (rbcL), RNA polymerase C (rpoC1), RNA polymerase B (rpoB), acetyl-CoA carboxylase-D (accD), maturase K (matK); and the intergenic regions ATP synthase subunit b-ATP synthase subunit c (atpF-atpH), Photosystem Ⅱ reaction center protein K-Photosystem Ⅱ reaction center protein I (psbK-psbI), the intergenic spacer region of psbA gene, and trnH gene (trnH-psbA). PCR was performed in 0.2 μL Eppendorf tubes inside of a Mini Thermal Cycler (Applied Biosystems 9700, USA) as per conditions optimized for each loci (Table 1). PCR products were resolved on agarose gels and purified using a QIAquick Gel Extraction Kit (Qiagen, USA). The purified PCR products were sequenced on an ABI 3730xl instrument (Applied Biosystems, USA) from Macrogen (South Korea) using the same set of primers as defined for PCR amplifications. Both forward and reverse primers were used for sequencing. The consensus sequences were generated in Geneious Pro (5.6.7) (Kearse et al., 2012).
2.3. Sequence analysisTo discriminate Kaempferia from Marantaceae species using barcodes, the genuine and adulterated sample were aligned and their sequence variation was quantified. The neighbor-joining (NJ) method was used to test phylogenetic relationships in MEGA 6.0 using K2P distances (2000 bootstrap replicates). These analyses were performed separately for matK, rbcL, rpoB, rpoC1, accD, atpF-atpH, trnH-psbA, and psbK-psbI; the DNA sequences were also concatenated in a combined analysis. All sequences generated from Kaempferia and the adulterant species were deposited in the NCBI database. We also used BLASTn searches to confirm our results.
3. Results and discussionTo test which loci are the most effective barcodes for discriminating Kaempferia from contaminants from the Marantaceae family, we assessed eight barcodes on six species (K. galanga, Kaempferia angustifolia, Kaempferia elegans, K. rotunda, C. bachemiana and Maranta leuconaria). These analyses produced 56 DNA sequences (Supplementary Table 1). Sequence characteristics of the eight plastid loci are listed in Table 2. Amplicon size varied from 400 bp in accD to 769 bp in matK, whereas in atpF-atpH, trnH-psbA and psbK-psbI, it was 750, 775, and 564 bp, respectively. The aligned length varied from 387 bp for accD to 769 bp for matK. Variable sites ranged from 1.15% for trnH-psbA and rpoC1 to 16.36% for atpF-atpH. When sequence length and number of conserved sites were taken into consideration, the most conserved sequences (above 94.00%) among all loci analyzed were trnH-psbA, rpoC1 and rbcL. atpF-atpH showed the highest nucleotide variation (16.36%), followed by psbK-psbI (14.18%), matK (13.91%) and accD (11.63%). Parsimony informative sites varied from 0.53% (rbcL) to 13.51% (atpF-atpH). The highest number of singleton sites was for psbK-psbI (4.79%), whereas trnH–psbA showed an absence of singleton sites.
| Amplicon size (bp) | Aligned length (bp) | Conserved sites | No of variable sites | Parsimony informative sites | Singleton sites | |
| accD | 400 | 387 | 342 | 45 | 31 | 14 |
| matK | 769 | 769 | 662 | 107 | 87 | 20 |
| rbcL | 600 | 566 | 551 | 14 | 3 | 11 |
| rpoB | 425 | 405 | 376 | 29 | 22 | 7 |
| rpoC1 | 525 | 534 | 505 | 29 | 21 | 8 |
| atpF-atpH | 750 | 703 | 585 | 115 | 95 | 20 |
| trnH -psbA | 775 | 695 | 687 | 8 | 8 | 0 |
| psbK-psbI | 564 | 564 | 456 | 80 | 53 | 27 |
| matK + rbcL | 1335 | 1209 | 126 | 90 | 36 | |
| rbcL + trnH-psbA | 1261 | 1238 | 22 | 11 | 11 | |
| matK + psbK-psbI | 1334 | 1109 | 189 | 147 | 42 | |
| matK + rbcL + rpoC1 | 1869 | 1715 | 154 | 111 | 43 | |
| matK + rbcL + rpoB | 1740 | 1582 | 158 | 112 | 46 | |
| matK + rbcL + accD | 1722 | 1551 | 171 | 121 | 50 | |
| matK + rbcL + atpF-atpH | 2038 | 1794 | 241 | 185 | 56 | |
| matK + rbcL + psbK-psbI | 1899 | 1665 | 206 | 146 | 60 | |
| matK + rbcL + trnH-psbA | 2030 | 1896 | 134 | 98 | 36 | |
| Eight plastid targets | 5113 | 3879 | 710 | 316 | 391 |
Individual barcoding loci (except for matK) were not effective at discriminating Kaempferia species from their adulterants (Supplementary Figs. 1–2). This finding is consistent with previous studies in which single-locus DNA barcodes from plastids were ineffective at discriminating plant species in Araucaria, Solidago, and Quercus (Fazekas et al., 2008, Hollingsworth, 2008, Piredda et al., 2011, Kress and Erickson, 2007). The ineffectiveness of these seven barcode loci (except matK) is likely due to hybridization events and the recent divergence of plant species within these families (Ley et al., 2014, Chen et al., 2015, Vinitha et al., 2014). Furthermore, research on molecular phylogeny has indicated that Kaempferia is a polyphyletic clade (Techaprasan et al., 2010), whereas species belonging to the Marantaceae family form a monophyletic clade, with Cannaceae as a sister group (Suksathan et al., 2009). In addition, hybridization and polyploidization have been shown to occur in the genus Kaempferia (Basak et al., 2018, Nopporncharoenkul et al., 2017), whereas most of Marantaceae species are diploids (Sharma and Mukhopadhyay, 1984). Our previous investigation on genome size of the studied Kaempferia species showed that K. elegans, K. angustifolia, and K. galanga are diploid, tetraploid, and pentaploid, respectively (Basak et al., 2018). The Marantaceae species in this study, however, are both diploid (data not shown). The divergence in ancestral relationship, along with difference in the ploidy level between Marantaceae and Kaempferia complex, may explain why individual barcodes failed to segregate Kaempferia and adulterants.
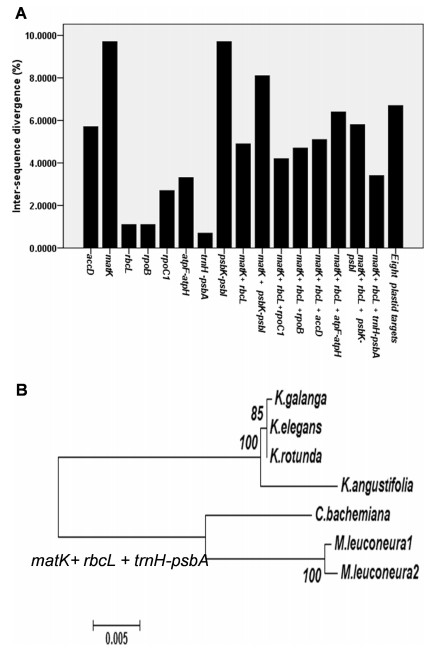
In phytogeographical studies of land plants, use of multi-locus barcodes is advised in cases where single-locus barcodes fail to discriminate species appropriately. To estimate the discriminatory power of eight barcoding loci and their combinations, we calculated interspecific variability using K2 parameters. The loci matK and psbK-psbI yielded higher interspecific variability (9.70%). The least variability was shown by trnH–psbA (0.70%), followed by rbcL and rpoB (1.10%) (Fig. 3). For barcoding land plants, previous studies have recommended using a two-locus system that consists of the matK + rbcL pair (CBOL Plant Working Group, 2009, Kress and Erickson, 2007). In our present investigation, the two-locus systems of matK + rbcL, matK + psbK-psbI, rbcL + trnH–psbA showed 126, 189 and 22 variable sites, respectively (Table 2). Among the two-locus barcode combinations, matK + rbcL provided the highest resolving power in species discrimination (Supplementary Fig. 3a). In contrast to previous studies (Kress and Erickson, 2007), we found that rbcL + trnH–psbA is not an effective barcode for reliable species discrimination (Supplementary Fig. 3c).
|
| Fig. 3 (a) Overall mean distance calculated using the K2 parameter between species belonging to the Kaempferia complex and their adulterants as a measure of the discriminatory power of the analysis. (b) Neighbor joining (NJ) tree for six species based on matK + rbcL + trnH-psbA sequence data. K. galanga is forming the tri-topological relationship with three other Kaempferia species in the dendrogram. The barcoding approach clearly distinguishes the Kaempferia complex from its adulterants showing the effectiveness of our study. Bootstrap percentages are shown above the branches which were obtained after 2000 replications. |
Although the two-locus barcode matK + rbcL provided adequate resolution to discriminate between plant species, we assessed multi-locus barcode combinations to increase the resolving power. We analyzed the effectiveness of the following concatenated barcode loci: matK + rbcL + accD, matK + rbcL + rpoB, matK + rbcL + rpoC1, matK + rbcL + atpF-atpH, matK + psbK-psbI, rbcL + rpoB + trnH-psbA(Supplementary Figs. 3a–3c; Supplementary Figs. 4a–4d; Supplementary Figs. 5a–5c). In all cases, the tri-topological relationship among the Kaempferia species were either absent or not supported by bootstrap values greater than 70%, making these phylogenetic relationships unsuitable for rapid molecular discrimination. According to bootstrap values and the tri-topological relationship among Kaempferia, the loci combinations that were most effective were concatenated matK, rbcL and trnH-psbA. BLASTn analyses confirmed that matK was successful at returning the right match. Computer analysis also confirmed that concatenated matK, rbcL and trnH-psbA resulted in higher resolution compared to matK alone (Supplementary Table 2). Thus, concatenation of three barcoding genes (matK, rbcL and trnH-psbA) was found to be effective for the discrimination of Kaempferia species from their adulterants.
4. ConclusionThis study presents a DNA barcoding method to rapidly identify Kaempferia species important in Ayurveda. We found that the matK locus can be used as a primary DNA barcode to discriminate between plants in the Kaempferia complex and their Marantaceae family adulterants. Furthermore, the rbcL + trnH-psbA combination of loci can serve as supplementary barcodes for the same purpose. These results strongly suggest that DNA barcoding is an effective approach to discriminate adulterant species from active species and ensure high quality Ayurvedic medicine.
The authors state that there is no conflict of interest.
SB and AMR thank MHRD for fellowship. LR thanks the Department of Biotechnology (DBT), Government of India, for funding the project by way of DBT Twinning Programme for NE (BT/33/NE/TBP/2010), MS Swaminathan Research Foundation for AFLP facility and Department of Biosciences and Bioengineering, IIT Guwahati, for providing all necessary infrastructural support.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.pld.2019.04.003.
Adyanthaya A., Ismail S., Sreelakshmi N., 2016. Indian traditional medicinal herbs against dental caries e an unsung past to a bright future. Saudi J. Oral Dental Res, 1(1): 1-6. |
Barrett C.F., Specht C.D., Leebens-Mack J., Stevenson D.W., Zomlefer W.B., Davis J.I., 2014. Resolving ancient radiations:can complete plastid gene sets elucidate deep relationships among the tropical gingers (Zingiberales)? Ann. Bot, 113: 119-133. DOI:10.1093/aob/mct264 |
Basak S., Krishnamurthy H., Rangan L., 2018. Genome size variation among 3 selected genera of Zingiberoideae. Meta Gene, 15: 42-49. DOI:10.1016/j.mgene.2017.11.003 |
Carlsen M.M., Fer T., Schmickl R., Leong-Skornickova J., Newman M., Kress W.J., 2018. Resolving the rapid plant radiation of early diverging lineages in the tropical Zingiberales:pushing the limits of genomic data. Mol. Phylogenet. Evol, 128: 55-68. DOI:10.1016/j.ympev.2018.07.020 |
CBOL Plant Working Group, 2009. A DNA barcode for land plants. Proc. Natl. Acad.Sci. Unit. States Am, 106: 12794-12797. DOI:10.1073/pnas.0905845106 |
Chen J., Zhao J., Erickson D.L., Xia N., Kress W.J., 2015. Testing DNA barcodes in closely related species of Curcuma (Zingiberaceae) from Myanmar and China. Mol. Ecol. Resour, 15: 337-348. DOI:10.1111/1755-0998.12319 |
Fazekas A.J., Burgess K.S., Kesanakurti P.R., et al, 2008. Multiple multilocus DNA barcodes from the plastid genome discriminate plant species equally well. PLoS One, 3: 2802. DOI:10.1371/journal.pone.0002802 |
Hollingsworth P.M., 2008. DNA barcoding plants in biodiversity hot spots:progress and outstanding questions Ⅲ. Heredity, 101: 1-2. |
Kearse M., Moir R., Wilson A., et al, 2012. Geneious Basic:an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics, 28: 1647-1649. DOI:10.1093/bioinformatics/bts199 |
Kress W.J., Erickson D.L., 2007. A two-locus global DNA barcode for land plants, the coding rbcL gene complements the non-coding trnH-psbA spacer region. PLoS One, 2: 508. DOI:10.1371/journal.pone.0000508 |
Kumar N.N., Ruba K., 2018. Identification of adulterants by pharmacognostic evaluation in selected medicinal plants. World J. Pharmaceut. Med. Res, 4(2): 67-70. |
Kumar S., Kahlon T., Chaudhary S., 2011. A rapid screening foradulterants in olive oil using DNA barcodes. Food Chem, 127: 1335-1341. DOI:10.1016/j.foodchem.2011.01.094 |
Ley, A.C., Dauby, G., Köhler, J., Wypior, C., Röser, M., Hardy, O.J., 2014. Comparative phylogeography of eight herbs and lianas (Marantaceae) in central African rainforests. Front. Genet. 5, 403. https://doi.org/10.3389/fgene.2014.00403.
|
Newmaster S.G., Grguic M., Shanmughanadhan D., Ramalingam S., 2013. DNA barcoding detects contamination and substitution in North American herbal products. BMC Med, 11: 222-235. DOI:10.1186/1741-7015-11-222 |
Nopporncharoenkul N., Chanmai J., Jenjittikul T., Anamthawat-Jónsson K., Soontornchainaksaeng P., 2017. Chromosome number variation and polyploidy in 19 Kaempferia (Zingiberaceae) taxa from Thailand and one species from Laos. J. Syst. Evol, 55: 466-476. DOI:10.1111/jse.12264 |
Parvathy V.A., Swetha V.P., Sheeja T.E., Leela N.K., Chompakam B., Sasikumar B., 2014. DNA barcoding to detect chilli adulteration in traded black pepper powder. Food Biotechnol, 28(1): 25-40. DOI:10.1080/08905436.2013.870078 |
Piredda R., Simeone M., Attimonelli M., et al, 2011. Prospects of barcoding the Italian wild dendroflora:oaks reveal severe limitations to tracking species identity. Mol. Ecol. Resour, 11: 72-83. DOI:10.1111/j.1755-0998.2010.02900.x |
Preetha T.S., Hemanthakumar A.S., Krishnan P.N., 2016. A comprehensive review of Kaempferia galanga L. (Zingiberaceae):a high sought medicinal plant in Tropical Asia. J. Med. Plant Stud, 4(3): 270-276. |
Rajashekhara N., Sharma P.P., 2010. A comparative study of efficacy of Tugaksheeree [Curcuma angustifolia Roxb. and Maranta arundinacea Linn.] in management of Amlapitta. Ayu, 31(4): 482-486. DOI:10.4103/0974-8520.82047 |
Sasikumar, B., Swetha, V.P., Parvathy, V.A., Sheeja, T.E., 2016. Advances in adulteration and authenticity testing of herbs and spices. In: Downey, G. (Ed.), Advances in Food Authenticity Testing. Woodhead Publishing Series in Food Science, Technology and Nutrition, pp. 585-624.
|
Septyanti C., Batubara I., Rafi M., 2016. HPLC fingerprint analysis combined with chemometrics for authentication of Kaempferia galanga from related species. Indones. J. Chem, 16(3): 308-314. |
Sereena K., Prakashkumar U., Remashree B., 2011. Histochemical and phytochemical markers for the authentication of ayurvedic raw drug hallakam (Kaempferia rotunda) and its marketed adulterant. Int. J. Pharm. Sci. Res, 2(11): 2952-2958. |
Sharma, A.K., Mukhopadhyay, S., 1984. Feulgen microspectrophotometric estimation of nuclear DNA of species and varieties of three different genera of marantaceae, 93 (3), 337-347.
|
Sivarajan, V.V., Balachandran, I., 1994. Ayurvedic Drugs and Their Plant Sources.Oxford and IBH Publishing Co. Pvt. Ltd., New Delhi.
|
Stoeckle M.Y., Gamble C.C., Kirpekar R., et al, 2011. Commercial teas highlight plant DNA barcode identification successes and obstacles. Sci. Rep, 1: 42-49. DOI:10.1038/srep00042 |
Suksathan P., Gustafsson M.H., Borchsenius F., 2009. Phylogeny and generic delimitation of Asian Marantaceae. Bot. J. Linn. Soc, 159: 381-395. DOI:10.1111/j.1095-8339.2009.00949.x |
Techaprasan J., Klinbunga S., Ngamriabsaku C., Jenjittikul T., 2010. Genetic variation of Kaempferia (Zingiberaceae) in Thailand based on chloroplast DNA (psbA-trnH and petA-psbJ) sequences. Genet. Mol. Res, 9(4): 1957-1973. DOI:10.4238/vol9-4gmr873 |
Vinitha M.R., Kumar U.S., Aishwarya K., Sabu M., Thomas G., 2014. Prospects for discriminating Zingiberaceae species in India using DNA barcodes. J. Integr.Plant Biol, 56: 760-773. DOI:10.1111/jipb.12189 |



