扩展功能
文章信息
- 张曦, 翟丽, 孙运艳, 杨伟, 高艳章, 雷鸣, 潘玉卿
- ZHANG Xi, ZHAI Li, SUN Yunyan, YANG Wei, GAO Yanzhang, LEI Ming, PAN Yuqing
- 基于高通量芯片对小儿急性髓系白血病的生物信息学分析
- Bioinformatics analysis of pediatric acute myeloid leukemia based on high-throughput microarray
- 吉林大学学报(医学版), 2020, 46(05): 1036-1042
- Journal of Jilin University (Medicine Edition), 2020, 46(05): 1036-1042
- 10.13481/j.1671-587x.20200522
-
文章历史
- 收稿日期: 2019-12-27
2. 云南省肿瘤医院·昆明医科大学第三附属医院血液科, 云南 昆明 650118;
3. 昆明医科大学第一附属医院检验科 云南省实验诊断研究所 云南省检验医学重点实验室, 云南 昆明 650032
2. Department of Hematology, Caner Hospital of Yunnan Province, Third Affiliated Hospital, Kunming Medical University, Kunming 650118, China;
3. Department of Clinical Laboratory, First Affiliated Hospital, Kunming Medical University, Yunnan Institute of Experimental Diagnosis, Yunnan Key Laboratory of Laboratory Medicine, Kunming 650032, China
小儿急性髓系白血病(acute myeloid leukemia,AML)约占儿童白血病总数的20% [1-2]。近年来,由于白血病诊疗方法和支持治疗的改进,AML患儿的长期生存率有了很大提高。虽然小儿AML的预后明显优于20岁以上的成人[3-5],但由于其化疗耐药、缓解率低和缓解期感染等因素阻碍了治愈率的提高[6],尤其是不明原因发热、体质量减轻、肺部浸润伴呼吸困难和肾功能衰竭等并发症在治疗过程中时有发生[7]。即使在发达国家,对复发难治性小儿白血病的治疗效果仍不乐观[8]。目前国内外对于小儿AML的治疗主要以放、化疗及造血干细胞移植为主,但对于其发病机制的报道相对较少,故探究小儿AML的分子机制并制定精准的治疗策略对于患儿的生存及预后尤为重要。迄今为止,白血病的病因尚不明确,其仍然是儿童和青少年癌症死亡的主要原因。本研究利用生物信息学分析手段对小儿AML患儿基因芯片进行分析,以期发现与该病相关的生物标志物,为揭示小儿AML发生的分子机制提供理论依据。
1 资料与方法 1.1 小儿髓系白血病基因芯片数据信息获取从GEO数据库(http://www.ncbi.nlm.nih.gov/geo)下载相关的基因表达矩阵,以“pediatric acute myeloid leukemia”为关键词筛选相关数据集。筛选条件:①mRNA表达谱数据集;②以骨髓为样本;③以正常骨髓为对照。选择基于GPL8300平台的GSE2191数据集(GeneChip Human Genome U95Av2 oligonucleotide microarray),包括54例小儿AML患儿(男性33例,女性21例,年龄0 d~15岁)和4名正常骨髓样本(正常人)。此外,本研究不包括任何对人体细胞或动物进行的实验。
1.2 基因芯片的处理及差异表达基因(differentially expressed genes,DEGs)的筛选采用GEO数据库自带的GEO2R在线工具对AML患儿及正常对照组间的DEGs进行筛选[9],筛选条件:错误发现率(false discovery rate,FDR) < 0.05且|log2 fold change(FC)| ≥ 2。以logFC ≥ 2为上调基因,logFC ≤-2为下调基因。下载MINiML平台文件,采用R语言的ggplot包绘制DEGs的火山图,pheatmap包绘制差异较大的前50个DEGs的热点图。
1.3 DEGs的基因本体功能注释(GO)富集和京都基因与基因组百科全书(KEGG)通路分析利用DAVID 6.8数据库分析GO富集和KEGG通路富集情况[10]。GO分析中以FDR < 0.05、基因数(gene counts) > 10作为筛选条件。KEGG分析中以P < 0.05作为筛选条件。采用R语言的GOplot包对GO富集实现可视化,并获得分子功能(molecular function,MF)、生物学过程(biological process,BP)和细胞成分(cellular component,CC)等注释分析。
1.4 蛋白互作网络(PPI)网络构建与核心基因(Hub gene)筛选采用SRTING数据库(https//stringdb.org)对本次研究中得到的DEGs构建PPI网络[11],并将得到的数据下载并导入Cytoscape软件(www.cytoscape.org/)进行可视化并去除游离蛋白质节点后,行CytoHubba插件计算每个节点的连接度(Degree),将连接度按照降序排列,并获得排名前20位的Hub基因[12]。
1.5 Hub基因参与疾病谱及相关转录因子分析将前5位的Hub基因通过生物技术信息基因云(gene-cloud of biotechnology information,GCBI)在线数据库进行分析,并采用Cytoscape中的iRegulon插件对20个Hub基因的相关转录因子进行预测,筛选条件为标准化富集分数(normalized enrichment score,NES) > 3[13]。
2 结果 2.1 AML患儿DEGs的筛选运用GEO2R功能进行DEGs识别,按照上述的DEGs筛选条件得到600个DEGs,其中407个基因上调,193个基因下调。数据集中的上调和下调基因如图 1A(插页七)所示。前50个DEGs如图 1B(插页七)所示。
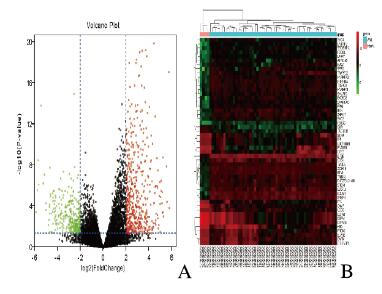
|
| A: Red represented the up-regulated genes, green represented the down-regulated gene,and black was the gene with no significant difference; B: Genes up-regulated were in red,genes down-regulatedwere in green, and genes without any significant difference were in black. The differences were set as corrected P value<0. 05 and log |FC|≥2. 图 1 小儿AML患者与正常对照样本DEGs的火山图(A)和前50个DEGs的热点图(B) Fig. 1 Volcano plots(A) and heatmap (B) of top 50 DEGs of AMIL patients and normal controls |
|
|
将DEGs上传到DAVID数据库,获得GO功能富集和KEGG通路分析。GO分析结果显示:DEGs主要富集在细胞成分中,包括核浆、细胞质、核膜和核斑点;在分子功能层面,DEGs主要涉及蛋白质及RNA的poly(A)尾结合;在生物学过程层面,DEGs主要涉及白细胞迁移。见图 2(插页七)和表 1。KEGG通路富集结果显示:DEGs主要富集在肿瘤坏死因子、细胞因子受体相互作用和Jak-STAT信号通路中。见表 2。
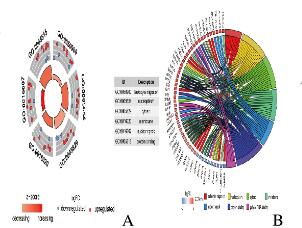
|
| A: The outer circle repre ese ented the expression of DEGs in each enriched GO term,red dots on each GO term indicated up-regulated genes and blue dots indicated down regulated genes.The inner circle indicated the significance of GO terms; B: The circle indicated the top 7 functional clusters in their gene ontology terms. 图 2 DEGs的GO富集分析 Fig. 2 GO enrichment analysis of DEGs |
|
|
| Category | Term | Description | Count | FDR |
| BP | GO:0050900 | Leukocyte migration | 16 | 1.25E-02 |
| CC | GO:0005829 | Cytosol | 151 | 1.99E-05 |
| CC | GO:0005654 | Nucleoplasm | 136 | 1.88E-06 |
| CC | GO:0016020 | Membrane | 108 | 1.40E-04 |
| CC | GO:0016607 | Nuclear speck | 21 | 3.20E-03 |
| MF | GO:0044822 | Poly(A) RNA binding | 344 | 3.72E-06 |
| MF | GO:0005515 | Protein binding | 198 | 1.58E-06 |
| E:Scientific notation. | ||||
| Category | Term | Discription | Count | P | |
| KEGG_PATHWAY | hsa04668 | TNF signaling pathway | 13 | 2.67E-04 | |
| KEGG_PATHWAY | hsa04060 | Cytokine-cytokine receptor interaction | 19 | 1.56E-03 | |
| KEGG_PATHWAY | hsa04630 | Jak-STAT signaling pathway | 13 | 3.87E-03 | |
| E:Scientific notation. | |||||
利用STRING数据库构建PPI网络,网络共涉及551个节点和773个连接。计算每个蛋白质节点的连接度,结果表明:PPI网络的最大连接度为24,最小连接度为16(图 3,见插页七)。排名前20位的Hub基因分别是甲酰肽受体2(formyl peptide receptor 2,FPR2)、磷酸肌醇3激酶调节亚单位1(phosphoinositide-3-kinase regulatory subunit 1,PIK3R1)、E1A结合蛋白p300(E1A binding protein p300,EP300)、热休克蛋白90α家族AA1(heat shock protein 90 alpha family AA1,HSP90AA1)、NRAS原癌基因(NRAS proto-oncogene,NRAS)、精氨酸酶1(arginase 1,ARG1)、磷脂酰肌醇4,5-二磷酸3-激酶催化亚单位α(phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit alpha,PIK3CA)、前血小板碱性蛋白(pro-platelet basic protein,PPBP)、CD59分子(CD59 molecule,CD59)、细胞骨架相关蛋白4(cytoskeleton associated protein 4,CKAP4)、乙酰辅酶a酰基转移酶1(acetyl-CoA acyltransferase 1,ACAA1)、抗菌肽(cathelicidin antimicrobial peptide,CAMP)、肽聚糖识别蛋白1(peptidoglycan recognition protein 1,PGLYRP1)、磷脂酶γ-1(phospholipase C gamma 1,PLCG1)、基质金属肽酶8(matrix metallopeptidase 8,MMP8)、乳糖转铁蛋白(lactotransferrin,LTF)、转氨酶1(transcobalamin 1,TCN1)、嗅介蛋白4(olfactomedin 4,OLFM4)、结合珠蛋白(haptoglobin,HP)和富半胱氨酸分泌蛋白3(cysteine rich secretory protein 3,CRISP3)。见表 3。
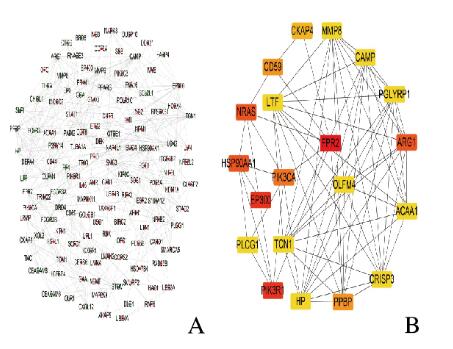
|
| A: Green nodes represented down-regulated genes,and red nodes represented up-regulated genes; B: Hub genes analyzed by cytoHubba in Cytoscape software. 图 3 DEGs的PPI网络构建和Hub基因筛选 Fig. 3 PPI network construction and Hub gene screening of DEGs |
|
|
| Gene | Gene description | Degree |
| FPR2 | Formyl peptide receptor 2 | 24 |
| PIK3R1 | Phosphoinositide-3-kinase regulatory subunit 1 | 23 |
| EP300 | E1A binding protein p300 | 23 |
| HSP90AA1 | Heat shock protein 90 alpha family class A member 1 | 22 |
| NRAS | NRAS proto-oncogene, GTPase | 22 |
| ARG1 | Arginase 1 | 22 |
| PIK3CA | Phosphatidylinositol-4, 5-bisphosphate 3-kinase catalytic subunit alpha | 20 |
| PPBP | Pro-platelet basic protein | 19 |
| CD59 | CD59 molecule | 19 |
| CKAP4 | Cytoskeleton associated protein 4 | 17 |
| ACAA1 | Acetyl-CoA acyltransferase 1 | 16 |
| CAMP | Cathelicidin antimicrobial peptide | 16 |
| PGLYRP1 | Peptidoglycan recognition protein 1 | 16 |
| PLCG1 | Phospholipase C gamma 1 | 16 |
| MMP8 | Matrix metallopeptidase 8 | 16 |
| LTF | Lactotransferrin | 16 |
| TCN1 | Transcobalamin 1 | 16 |
| OLFM4 | Olfactomedin 4 | 16 |
| HP | Haptoglobin | 16 |
| CRISP3 | Cysteine rich secretory protein 3 | 16 |
GCBI分析结果显示:EP300、HSP90AA1和NRAS共3个Hub基因参与了小儿AML的发生发展(图 4);通过Cytoscape中的iRegulon插件预测DEGs中排名前20个Hub基因的转录因子,结果显示:共有55个转录因子调节DEGs,排名前15位转录因子结果见表 4。部分转录因子的作用关系见图 5。
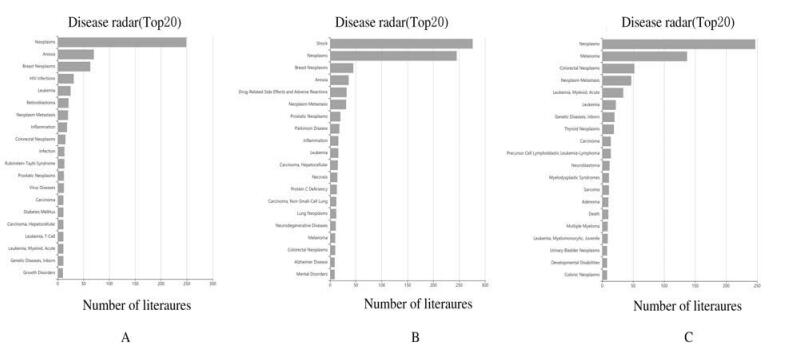
|
| 图 4 Ep300(A)、HSP90AA1(B)和NRAS(C)的相关疾病谱 Fig. 4 Disease spectrum of EP300(A), HSP90AA1(B), and NRAS(C) |
|
|
| TF | NES | Target gene | Transcription negulatory element |
| TP63 | 7.482 | 11 | 1 |
| NFE2L1 | 6.442 | 10 | 4 |
| TBX19 | 6.210 | 10 | 3 |
| CBFB | 5.813 | 6 | 5 |
| SMARCA4 | 5.773 | 3 | 1 |
| POU1F1 | 5.647 | 8 | 4 |
| HSF1 | 5.524 | 7 | 5 |
| USP39 | 5.406 | 13 | 6 |
| ING4 | 5.401 | 6 | 2 |
| TFCP2 | 5.378 | 3 | 2 |
| SMAD1 | 5.335 | 6 | 9 |
| TAF1 | 5.183 | 3 | 1 |
| JUN | 5.119 | 3 | 1 |
| SMARCB1 | 5.076 | 3 | 1 |
| KMD4B | 5.075 | 3 | 2 |
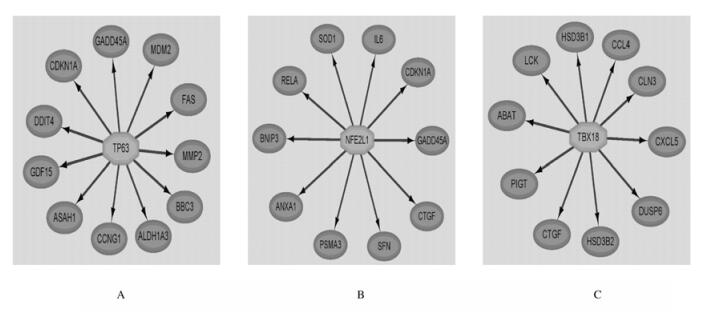
|
| A:TP63;B:NFE2L1;C:TBX19. 图 5 转录因子的靶基因作用关系图 Fig. 5 Interaction networks of target genes for transcription factors |
|
|
1~14岁儿童所患肿瘤中近1/3是白血病,其中3/4为急性淋巴细胞性白血病(acute lymphoblastic leukaemia,ALL)[14]。虽然小儿AML所占比例不大,但如果不经任何治疗,AML会在发病初期对儿童造成极大的危害[15]。在白血病发生发展过程中参与造血调控的基因常发生突变,导致造血细胞分化缺陷。而每种类型的白血病都有不同的基因突变。临床上,白血病治疗和生存率主要取决于基因突变的类型和诊断分期[16]。为此,本研究从GEO数据库下载小儿AML的基因表达矩阵,筛选出可能参与小儿AML的核心基因,旨在为小儿AML的发病机制、诊断及防治提供新的理论依据和研究视角。
本研究采用STRING数据库构建PPI网络后,通过Cytoscape软件对网络进行可视化。利用CytoHubba插件计算每个蛋白质节点的连接度,共筛选出Ep300、HSP90AA1和NRAS等20个与小儿AML相关的Hub基因,对上述基因进行文献检索后发现:对维甲酸处理的急性早幼粒细胞能够显著增加细胞分化时膜表面的抗炎因子Annexin A1及其受体分子FPR2的表达[17]。对诱导化疗失败的AML患儿进行全基因组DNA、转录组RNA和miRNA测序结果显示:PIK3R1参与小儿AML早期化疗耐药的遗传机制[18],而PIK3R1作为白血病的超级增强子抑制了细胞凋亡并促进其增殖[19]。Ep300基因表达下调并作为抑癌因子参与小儿AML的进程[20]。在造血干细胞和祖细胞内,由于Ep300基因的缺失直接增强了MAPK及JAK/STAT等细胞因子的信号通路,Ep300基因甚至能够阻断造血异常综合征(human myelodysplastic syndrome,MDS)向AML的转变[21]。HSP90AA1基因作为重要的分子伴侣参与细胞增殖、存活和适应的信号传导途径。抑制HSP90AA1基因的表达活性后,能将其作为研究AML的重要分子靶标[22]。单核苷酸变异分析[23]显示:NRAS在初诊为急性早幼粒细胞白血病且维甲酸基因发生重排的患儿体内发生突变并参与了白血病的发病机制。NRAS基因作为热点区域突变基因亚群,在小儿AML的分子流行病学和生物学研究中具有重要意义[24]。通过维甲酸及1,25二羟基维生素D3的诱导,髓系白血病细胞能够成功向M2型巨核细胞进行分化,此时其表面标记分子ARG1的表达上调,提示其能够成为AML化疗成功与否的标志物[25]。
此外,本研究还应用Cytoscape中的iRegulon插件预测Hub基因的转录因子,并分析转录因子所调节的DEGs,结果显示:与20个Hub基因相关的转录因子有55个,其中有部分转录因子在白血病中的作用已经得到证实。TP63与长链非编码RNA rs55829688发生相互作用,加剧了AML患者的骨髓抑制,并影响该病的预后[26]。在多发性骨髓瘤中,长链非编码RNA MALAT1的拮抗作用下调了蛋白酶体亚单位基因的2个主要转录激活因子(NFE2L1和NRF2),导致胰蛋白酶、糜蛋白酶和细胞凋亡蛋白酶活性降低,并导致多泛素蛋白的累积[27]。
综上所述,本研究采用GEO数据库中的小儿AML芯片信息,利用一系列的生物信息学手段,探寻小儿AML相对精准的预后标志物,为其诊治提供更为有效的数据支持和理论依据。
| [1] |
CREUTZIG U, KUTNY M A, BARR R, et al. Acute myelogenous leukemia in adolescents and young adults[J]. Pediatr Blood Cancer, 2018, 65(9): e27089. DOI:10.1002/pbc.27089 |
| [2] |
KAHN J M, KEEGAN T H, TAO L, et al. Racial disparities in the survival of American children, adolescents, and young adults with acute lymphoblastic leukemia, acute myelogenous leukemia, and Hodgkin lymphoma[J]. Cancer, 2016, 122(17): 2723-2730. DOI:10.1002/cncr.30089 |
| [3] |
DUGGAN M A, ANDERSON W F, ALTEKRUSE S, et al. The Surveillance, epidemiology, and end results (SEER) program and pathology:Toward strengthening the critical relationship[J]. Am J Surg Pathol, 2016, 40(12): e94-e102. |
| [4] |
M RICKE A, ZIMMERMANN M, REITER A, et al. Long-term results of five consecutive trials in childhood acute lymphoblastic leukemia performed by the ALL-BFM study group from 1981 to 2000[J]. Leukemia, 2010, 24(2): 265-284. DOI:10.1038/leu.2009.257 |
| [5] |
RASCHE M, ZIMMERMANN M, BORSCHEL L, et al. Successes and challenges in the treatment of pediatric acute myeloid leukemia:a retrospective analysis of the AML-BFM trials from 1987 to 2012[J]. Leukemia, 2018, 32(10): 2167-2177. DOI:10.1038/s41375-018-0071-7 |
| [6] |
LANDRIGAN P J. Childhood leukemias[J]. N Engl J Med, 1995, 333(19): 1286. DOI:10.1056/NEJM199511093331912 |
| [7] |
SANZ M A, MONTESINOS P. How we prevent and treat differentiation syndrome in patients with acute promyelocytic leukemia[J]. Blood, 2014, 123(18): 2777-2782. DOI:10.1182/blood-2013-10-512640 |
| [8] |
HUDSON M M, LINK M P, SIMONE J V. Milestones in the curability of pediatric cancers[J]. J Clin Oncol, 2014, 32(23): 2391-2397. DOI:10.1200/JCO.2014.55.6571 |
| [9] |
DAVIS S, MELTZER P S. GEOquery:a bridge between the gene expression omnibus (GEO) and BioConductor[J]. Bioinformatics, 2007, 23(14): 1846-1847. DOI:10.1093/bioinformatics/btm254 |
| [10] |
HUANG D W, SHERMAN B T, LEMPICKI R A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources[J]. Nat Protoc, 2009, 4(1): 44-57. DOI:10.1038/nprot.2008.211 |
| [11] |
SZKLARCZYK D, GABLE A L, LYON D, et al. STRING v11:protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets[J]. Nucleic Acids Res, 2019, 47(D1): D607-D613. DOI:10.1093/nar/gky1131 |
| [12] |
SHANNON P, MARKIEL A, OZIER O, et al. Cytoscape:a software environment for integrated models of biomolecular interaction networks[J]. Genome Res, 2003, 13(11): 2498-2504. DOI:10.1101/gr.1239303 |
| [13] |
JANKY R, VERFAILLIE A, IMRICHOV H, et al. iRegulon:from a gene list to a gene regulatory network using large motif and track collections[J]. PLoS Comput Biol, 2014, 10(7): e1003731. DOI:10.1371/journal.pcbi.1003731 |
| [14] |
SIEGEL R, DESANTIS C, VIRGO K, et al. Cancer treatment and survivorship statistics, 2012[J]. CA Cancer J Clin, 2012, 62(4): 220-241. DOI:10.3322/caac.21149 |
| [15] |
KAYSER S, LEVIS M J. Advances in targeted therapy for acute myeloid leukaemia[J]. Br J Haematol, 2018, 180(4): 484-500. DOI:10.1111/bjh.15032 |
| [16] |
NTZIACHRISTOS P, MULLENDERS J, TRIMARCHI T, et al. Mechanisms of epigenetic regulation of leukemia onset and progression[J]. Adv Immunol, 2013, 117: 1-38. DOI:10.1016/B978-0-12-410524-9.00001-3 |
| [17] |
TSAI W H, CHIEN H Y, SHIH C H, et al. Annexin A1 mediates the anti-inflammatory effects during the granulocytic differentiation process in all-trans retinoic acid-treated acute promyelocytic leukemic cells[J]. J Cell Physiol, 2012, 227(11): 3661-3669. DOI:10.1002/jcp.24073 |
| [18] |
MCNEER N A, PHILIP J, GEIGER H, et al. Genetic mechanisms of primary chemotherapy resistance in pediatric acute myeloid leukemia[J]. Leukemia, 2019, 33(8): 1934-1943. DOI:10.1038/s41375-019-0402-3 |
| [19] |
WONG R W J, NGOC P C T, LEONG W Z, et al. Enhancer profiling identifies critical cancer genes and characterizes cell identity in adult T-cell leukemia[J]. Blood, 2017, 130(21): 2326-2338. DOI:10.1182/blood-2017-06-792184 |
| [20] |
XIAO P F, TAO Y F, HU S Y, et al. mRNA expression profiling of histone modifying enzymes in pediatric acute monoblastic leukemia[J]. Pharmazie, 2017, 72(3): 177-186. |
| [21] |
CHENG G, LIU F, ASAI T, et al. Loss of p300 accelerates MDS-associated leukemogenesis[J]. Leukemia, 2017, 31(6): 1382-1390. DOI:10.1038/leu.2016.347 |
| [22] |
WALSBY E J, LAZENBY M, PEPPER C J, et al. The HSP90 inhibitor NVP-AUY922-AG inhibits the PI3K and IKK signalling pathways and synergizes with cytarabine in acute myeloid leukaemia cells[J]. Br J Haematol, 2013, 161(1): 57-67. DOI:10.1111/bjh.12215 |
| [23] |
ZHAO J, LIANG J W, XUE H L, et al. The genetics and clinical characteristics of children morphologically diagnosed as acute promyelocytic leukemia[J]. Leukemia, 2019, 33(6): 1387-1399. DOI:10.1038/s41375-018-0338-z |
| [24] |
ANDRADE F G, NORONHA E P, BRISSON G D, et al. Molecular characterization of pediatric acute myeloid leukemia:results of a multicentric study in Brazil[J]. Arch Med Res, 2016, 47(8): 656-667. DOI:10.1016/j.arcmed.2016.11.015 |
| [25] |
TAKAHASHI H, HATTA Y, IRIYAMA N, et al. Induced differentiation of human myeloid leukemia cells into M2 macrophages by combined treatment with retinoic acid and 1α, 25-dihydroxyvitamin D3[J]. PLoS One, 2014, 9(11): e113722. DOI:10.1371/journal.pone.0113722 |
| [26] |
YAN H, ZHANG D Y, LI X, et al. Long non-coding RNA GAS5 polymorphism predicts a poor prognosis of acute myeloid leukemia in Chinese patients via affecting hematopoietic reconstitution[J]. Leuk Lymphoma, 2017, 58(8): 1948-1957. DOI:10.1080/10428194.2016.1266626 |
| [27] |
AMODIO N, STAMATO M A, JULI G, et al. Drugging the lncRNA MALAT1 via LNA gapmeR ASO inhibits gene expression of proteasome subunits and triggers anti-multiple myeloma activity[J]. Leukemia, 2018, 32(9): 1948-1957. DOI:10.1038/s41375-018-0067-3 |
 2020, Vol. 46
2020, Vol. 46


