扩展功能
文章信息
- 郭玮玮, 秦悦, 杨海波, 米占虎
- GUO Weiwei, QIN Yue, YANG Haibo, MI Zhanhu
- LncRNA MALAT1通过miR-34c/SATB2轴对脂肪间充质干细胞成骨分化的促进作用
- Promotion effect of LncRNA MALAT1 on osteogenic differentiation of adipose-derived mesenchymal stem cells through miR-34c/SATB2 axis
- 吉林大学学报(医学版), 2020, 46(05): 963-971
- Journal of Jilin University (Medicine Edition), 2020, 46(05): 963-971
- 10.13481/j.1671-587x.20200511
-
文章历史
- 收稿日期: 2020-02-10
脂肪间充质干细胞(adipose-derived stem cells,ADSCs)具有自我更新和多向分化潜能,可用于再生或修复骨组织[1]。与骨髓来源的间充质干细胞比较,ADSCs组织来源更丰富,细胞增殖速度更快[2]。然而,尽管有大量研究ADSCs可分化为多种谱系,但ADSCs分化的分子机制仍不明了。长链非编码RNA(long non-coding RNA,LncRNA)和微小RNA(miRNA)在成骨细胞分化中起关键作用[3-4]。研究[5]表明:LncRNA-KCNQ1OT1通过激活Wnt/β-catenin通路促进成骨分化以减弱骨溶解。LncRNA H19的异常表达与成骨分化有关[6]。细胞实验[7]表明:LncRNA肺癌转移相关转录本1(metastasis-associated lung ademocarcinoma transcript 1,MALAT1)通过抑制miR-204的表达,促进成骨细胞特异性标志物碱性磷酸酶(ALP)、骨钙蛋白(OCN)和矿化的骨基质形成。研究[8]表明:miRNA参与调节成骨细胞和破骨细胞分化过程。miR-34c通过调节成骨细胞中特异AT序列结合蛋白2(special AT-rich sequence binding protein 2,SATB2)和Runt相关转录因子2(Runx2)等多个靶点,影响成骨细胞和破骨细胞在体内的骨稳态[9]。过表达SATB2可以促进乙醇诱导的骨坏死患者骨髓间充质干细胞(bone marrow mesenchyml stem cells,BMSCs)的成骨分化[10]。目前,LncRNA MALAT1在ADSCs成骨分化中的作用尚不清楚。本研究旨在探讨LncRNA MALAT1通过miR-34c/SATB2轴在ADSCs成骨分化中的作用,以期为明确LncRNA MALAT1调控ADSCs成骨分化的机制和骨损伤的治疗提供科学指导。
1 材料与方法 1.1 细胞、主要试剂和仪器原代人ADSCs购自美国ScienCell公司,HEK-293T细胞购自中国典型培养物保藏中心。DMEM培养基购自美国Hyclone公司,胎牛血清(FBS)购自美国Gibco公司,青链霉素混合液(100×)购自北京索莱宝科技有限公司,Lipofectamine 3000转染试剂购自美国Invitrogen公司,SYBRPremix Ex TaqTMⅡ试剂盒购自日本TaKaTa公司,BCA蛋白定量试剂盒购自北京全式金公司,lentiviral-pEF-1a/Puro-NC(Lenti-NC)、lentiviral-pEF-1a/Puro-MALAT1(Lenti-MALAT1)、lentiviral-pEF-1a/Puro- SATB2(Lenti-SATB2)、lentiviralpGLVU6/Puro-sh-NC(sh-NC)、lentiviralpGLVU6/Puro-sh-MALAT1(sh-MALAT1)、miR-NC和miR-34c mimic(miR-34c)均购自上海GenePharma公司,油红O和茜素红购自美国Sigma公司,FITC-CD44抗体、FITC-CD29抗体、PE-CD90抗体、PE-CD105抗体和PE-vWF抗体购自美国BD公司,Runx2抗体、OPN抗体和OCN抗体购自英国Abcam公司。倒置相差显微镜购自日本Nikon公司,实时荧光定量PCR仪购自瑞士Roche公司,微量分光光度计购自美国Thermo Scientific公司。
1.2 细胞培养ADSCs和HEK-293T细胞用含10% FBS、100 IU·mL-1青霉素和100 mg·L-1链霉素的DMEM培养基中,于37℃、5% CO2条件下培养。
1.3 成骨细胞分化诱导诱导ADSCs成骨分化:当ADSCs长至80%~90%密度时,将培养基替换为成骨细胞特异性诱导培养基(含10%FBS,10 mmol·L-1 β-甘油磷酸钠,0.1 μmol·L-1地塞米松和0.2 mmol·L-1抗坏血酸磷酸的低糖DMEM),每2 d更换1次培养基,于37℃、5%CO2条件下培养0、3、7、14和21 d。诱导ADSCs成脂分化:当ADSCs长至80%~90%密度时,将培养基替换为成脂细胞特异性诱导培养基(含10%FBS,10 mmol·L-1 β-甘油磷酸钠,1 μmol·L-1地塞米松,0.5 mmol·L-1 3-异丁基-1-甲基黄嘌呤,10 μmol·L-1胰岛素和200 μmol·L-1吲哚美辛的低糖DMEM),每2 d更换1次培养基,于37℃、5%CO2条件下培养14 d。
1.4 免疫荧光染色检测ADSCs中CD44、CD29、CD90、CD105和vWF表达ADSCs成骨诱导培养基培养21 d后,弃去细胞培养基,PBS缓冲液清洗后,4%多聚甲醛固定,0.1% Triton X-100穿透,10%脱脂奶粉封闭,FITC-CD44抗体(1︰200)、FITC-CD29抗体(1︰200)、PE-CD90抗体(1︰200)、PE-CD105抗体(1︰200)和PE-vWF抗体(1︰200)避光孵育2 h,DAPI染色,荧光显微镜下观察细胞发光情况。
1.5 油红O染色法检测细胞脂滴形成ADSCs弃去成脂诱导培养基,PBS缓冲液清洗后,4%多聚甲醛固定。PBS缓冲液清洗细胞后,0.3%油红O室温染色20 min。弃去染液,蒸馏水洗涤后,显微镜观察并拍照。
1.6 ADSCs中ALP水平检测ADSCs弃去成骨诱导培养基,根据BCIP/NBT碱性磷酸酯酶显色试剂盒说明书检测细胞中ALP水平,酶标仪405 nm处检测各组细胞吸光度(A)值,对ALP水平进行定量。计算公式:ALP水平=[(样品组A值-空白组A值)-(对照组A值-空白组A值)]/(对照组A值-空白组A值)。
1.7 RT-PCR法检测ADSCs中miR-34c表达和LncRNA MALAT1、SATB2、Runx2、OPN及OCN mRNA表达水平收集转染后的ADSCs,TRIzol法提取样品中的总RNA,分光光度计测定RNA浓度,逆转录合成cDNA。按照SYBRPremix Ex TaqTMⅡ试剂盒说明书进行RT-PCR法检测。反应条件:95℃、30 s;95℃、5 s,58℃、30 s,40个循环;95℃、15 s,58℃、30 s,95℃、15 s。以U6和GAPDH为内参,采用2-ΔΔCt法计算miR-34c表达水平和LncRNA MALAT1、SATB2、Runx2、OPN及OCN mRNA表达水平。引物序列见表 1。
| Primer | Primer sequence (5'-3') |
| miR-34c | F:ACACTCCAGCTGGGAGGCAGTGTAGTT- AGCT |
| R:TGGTGTCGTGGAGTCG | |
| LncRNA MALAT1 |
F:GGTAACGATGGTGTCGAGGTC |
| R:CCAGCATTACAGTTCTTGAACATG | |
| SATB2 | F:GCAGTTGGACGGCTCTCTT |
| R:CACCTTCCCAGCTTGATTATTCC | |
| Runx2 | F:GACGAGGCAAGAGTTTCACC |
| R:GGTTCCCGAGGTCCATCTAC | |
| OPN | F:ATCTCCTAGCCCCACAGAAT |
| R:CATCAGACTGGTGAGAATCATC | |
| OCN | F:AGGGCAGCGAGGTAGTGA |
| R:CCTGAAAGCCGATGTGGT | |
| GAPDH | F:AGACAGCCGCATCTTCTTGT |
| R:TGATGGCAACAATGTCCACT | |
| U6 | F:CGCTTCGGCAGCACATATACTAAAATT⁃ GGAAC |
| R:GCTTCACGAATTTGCGTGTCATCCTTGC |
成骨诱导后的ADSCs或293T细胞接种于6孔细胞培养板,每孔4×105个细胞,待细胞密度达到60%~70%时,按照Lipofectamine 3000说明书进行miR-34c和miR-NC的转染,终浓度均为60 nmol·L-1。37℃、5%CO2条件下培养48 h。ADSCs转染慢病毒时,将细胞培养液更换为含6 mg·L-1聚凝胺和感染复数(MOI)=20的lentiviral-pEF-1a/Puro-NC(Lenti-NC)、lentiviral-pEF-1a/Puro-MALAT1(Lenti-MALAT1)、lentiviral-pEF-1a/Puro-SATB2(Lenti- SATB2)、lentiviralpGLVU6/Puro-sh-NC(sh-NC)和lentiviralpGLVU6/Puro-sh-MALAT1(sh-MALAT1)慢病毒的新鲜培养基,37℃、5%CO2条件下培养48 h。
1.9 荧光素酶报告实验检测LncRNA MALAT1与miR-34c和miR-34c与SATB2之间的靶向结合作用miRcode(http://www.miRcode.org/)预测MALAT1与miR-34c的靶向结合位点,TargetScan 7.1(http://www.TargetScan.org/vert_/)预测miR-34c和SATB2之间的靶向结合位点。针对LncRNA MALAT1 3'-UTR端和SATB2 3'-UTR端序列构建野生型(WT)和突变型(MUT)报告基因质粒。参照“1.8”步骤,将MALAT1-WT和MALAT1-MUT(或SATB2-WT和SATB2-MUT)分别与miR-NC和miR-34c共同转染至HEK-293T细胞中,48 h后检测荧光素酶活性。
1.10 茜素红S(ARS)染色检测细胞钙盐沉积弃去ADSCs成骨诱导培养基,PBS缓冲液清洗后,4%多聚甲醛固定。PBS缓冲液清洗细胞后,0.1%ARS染色液室温孵育20 min。弃去染液,蒸馏水洗涤。为了定量钙盐沉积,向细胞中加入含有10 mmol·L-1磷酸钠的10%氯化十六烷基吡啶,并用酶标仪检测各样品在562 nm处的A值。ARS水平=[(样品组A值-空白组A值)-(对照组A值-空白组A值)]/(对照组值-空白组A值)。
1.11 Western blotting法检测细胞中Runx2、OPN和OCN蛋白表达水平收集转染后的ADSCs或293T细胞,BCA法测定蛋白浓度,取30 μg蛋白进行SDS-PAGE凝胶电泳分离蛋白,并将其转至PVDF膜上。10%脱脂奶粉室温封闭3 h,加入Runx2(1︰1 500)、OPN(1︰1 000)、OCN(1︰2 000)和GAPDH(1︰2 000),4℃孵育过夜。HRP标记二抗(1︰2 500)室温孵育1 h后,ECL发光、曝光。Image J分析条带灰度值,以目的蛋白条带灰度值与GAPDH条带灰度值比值表示目的蛋白表达水平。
1.12 统计学分析采用SPSS 21.0统计软件进行统计学分析。各组细胞中荧光素酶活性,LncRNA MALAT1、miR-34、SATB2、Runx2、OPN和OCN表达水平,ALP和ARS水平均以x±s表示,2组间样本均数比较采用两独立样本t检验,多组间样本均数比较采用单因素方差分析。以P < 0.05为差异有统计学意义。
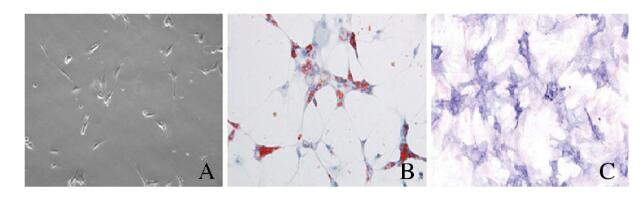
2 结果 2.1 ADSCs的特征和多能分化潜能的鉴定ADSCs中间充质干细胞标记物CD29、CD90、CD44和CD105呈阳性表达,而内皮细胞标记物vWF呈阴性表达(图 1A~F,见插页二);同时细胞具有长丝状结构(图 2A,见插页三),表明该细胞是ADSCs。油红O染色法和ALP染色检测结果显示:在成脂或成骨诱导条件下,ADSCs中有红色脂滴形成(图 2B,见插页三)和橘红色钙盐沉积(图 2C,见插页三)。

|
| A: CD29; B: CD90; C: CD44; D: CD105; E: vWF [labeled with FITC (green color) or PE (red color)]; F: Negative control. 图 1 免疫荧光染色检测ADSCs中CD29.CD90、CD44.CD105和vWF的表达(× 400) Fig. 1 Expressions of CD29, CD90, CD44, CD105 and vWF in ADSCs detected by immunofluorescence staining(× 400) |
|
|
|
| A: Typical cobblestone-like morphology of ADSCs; B: Oil red Ostaining; C: Alizarin red staining. 图 2 ADSCs多向分化潜能的鉴定(× 200) Fig. 2 Identification of multi- differentiation potential of ADSCs(× 200) |
|
|
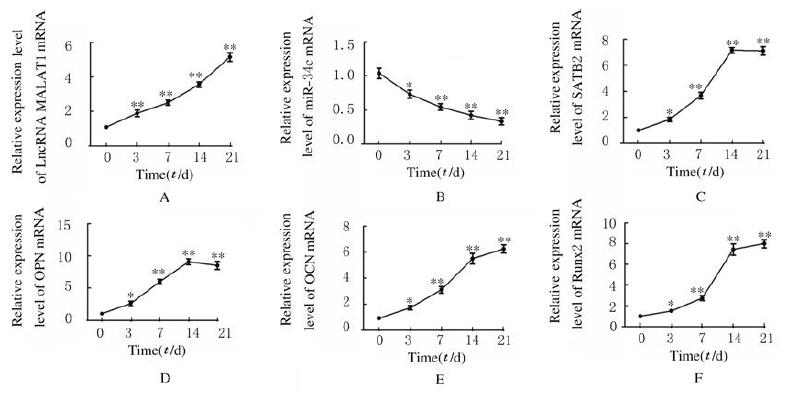
与第0天比较,ADSCs成骨诱导第3、7、14和21天后细胞中LncRNA MALAT1和SATB2及成骨标记物Runx2、OPN和OCN mRNA表达水平均明显升高(P < 0.05),miR-34c mRNA表达水平明显降低(P < 0.05)。见图 3。
|
| *P < 0.05, **P < 0.01 vs day 0. 图 3 成骨分化过程中ADSCs中LncRNA MALAT1(A)、miR-34c(B)、SATB2(C)、OPN(D)、OCN(E)和Runx2(F)mRNA表达水平 Fig. 3 Expression levels of LncRNA MALAT1 (A), miR-34c (B), SATB2 (C), OPN (D), OCN(E), and Runx2(F) mRNA in ADSCs during osteogenie differentiation |
|
|
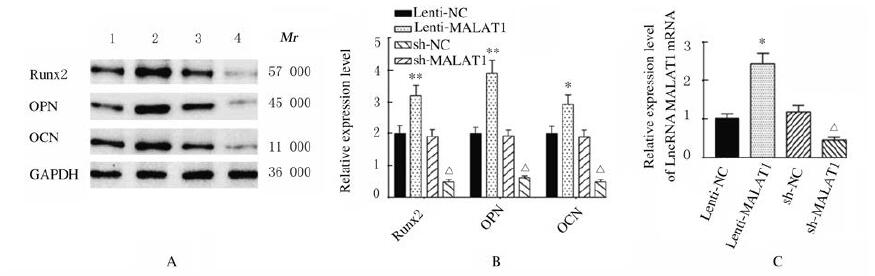
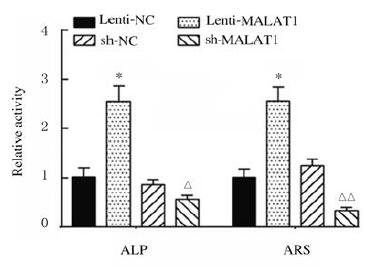
与Lenti-NC组比较,Lenti-MALAT1组ADSCs中Runx2、OPN和OCN蛋白表达水平明显升高(P < 0.01),见图 4A,LncRNA MALAT1蛋白表达水平明显升高(P < 0.01),见图 4B,ALP和ARS活水平明显升高(P < 0.01),见图 5(插页三)和图 6;与sh-NC组比较,sh-MALAT1组ADSCs中Runx2、OPN和OCN蛋白表达水平明显降低(P < 0.01),见图 4A,MALAT1蛋白表达水平明显降低(P < 0.01),见图 4B,ALP和ARS水平明显降低(P < 0.01),见图 5(插页三)和图 6。
|
| Lane 1: Lenti-NC group; Lane 2: Lenti-MALAT1 group; Lane 3: sh-NC group; Lane 4: sh-MALAT1 group. *P < 0.05, **P < 0.01 vs Lenti-NC group; △P < 0.01 vs sh-NC group. 图 4 各组ADSCs中LncRNA MALAT1、Runx2、OPN和OCN蛋白表达电泳图(A)和直条图(B, C) Fig. 4 Electrophoregram (A) and histogram (B, C) of expressions of MALAT1, Runx2, OPN and OCN proteins in ADSCs in various groups |
|
|

|
| 图 5 ALP和ARS染色检测ADSCs中ALP活性和钙盐沉积 Fig. 5 ALP activities and calcium salt deposition in ADSCs detected by ALP and ARS staining |
|
|
|
| *P < 0.01 vs Lenti-NC group; △P < 0.05, △△P < 0.01 vs sh-NC group. 图 6 各组ADSCs中ALP和ARS水平 Fig. 6 Levels of ALP and ARS in ADSCs in various groups |
|
|
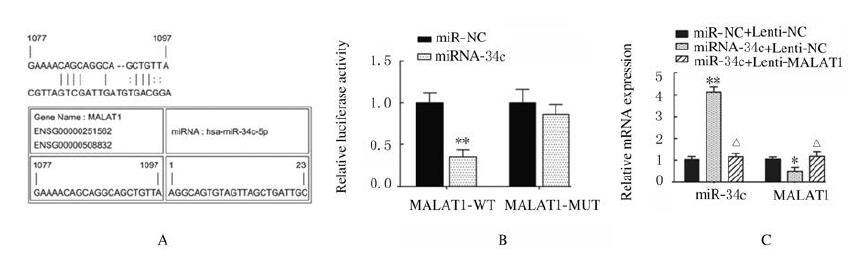
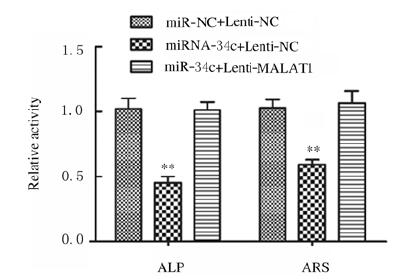
LncrRNA MALAT1与miR-34c结合位点见图 7A。荧光素酶报告实验结果显示:与miR-NC组比较,miR-34c转染后MALAT1-WT组荧光素酶活性明显降低(P < 0.01),MALAT1-MUT组荧光素酶活性差异无统计学意义(P > 0.05,见图 7B)。与miR-NC+Lenti-NC组比较,miR-34c+Lenti-NC组ADSCs中miR-34c表达水平明显升高(P < 0.01,见图 7C),LncRNA MALAT1表达水平明显降低(P < 0.05,见图 7C),Runx2、OPN和OCN蛋白表达水平明显降低(P < 0.01),见图 8,ALP和ARS水平明显降低(P < 0.01,见图 9(插页三)和图 10,miR-34c+Lenti-MALAT1组ADSCs中miR-34c、MALAT1表达水平及Runx2、OPN和OCN表达水平,ALP和ARS水平差异无统计学意义(P > 0.05)。
|
| B:*P < 0.05, **P < 0.01 vs miR-NC group; C:*P < 0.05, **P < 0.01vs miR-NC+Lenti-NC group; △P < 0.05, △△P < 0.01 vs miR-34c+Lenti-NC group. 图 7 miRcode、荧光素酶报告实验和RT-PCR法检测各组细胞中miR-34c和LncRNA MALAT1的靶向结合(A)及各组细胞中荧光素酶活性、miR-34c和MALAT1 mRNA表达水平(B,C) Fig. 7 Targeted binding of miR-34c and MALAT1 (A) and activities of luciferase expression levels of miR-34c and LncRNA MALAT1 in cells in various groups(B, C) detected by miRcode, luciferase report experiment and RT-PCR methods |
|
|

|
| Lane 1: miR-NC+Lenti-NC; Lane 2: miR-34c +Lenti-NC group; Lane 3: miR-34c+Lenti-MALAT1 group. *P < 0.01 vs miR-NC+Lenti-NC group. 图 8 Western blotting法检测共转染组ADSCs中Runx2、OPN和OCN蛋白表达电泳图(A)和直条图(B) Fig. 8 Electrophoregram(A) and histogram(B) of expressions of Runx2, OPN and OCN proteins in ADSCs in co-transfection groups detected by Western blotting method |
|
|

|
| 图 9 ALP和ARS染色检测共转染组ADSCs中ALP活性和钙盐沉积 Fig. 9 ALP activities and calcium salt deposition in ADSCs in co-transfection groups detected by ALP and ARS staining |
|
|
|
| *P < 0.05, **P < 0.01 vs miR-NC+Lenti-NC group. 图 10 共转染组ADSCs中ALP和ARS水平 Fig. 10 Levels of ALP and ARS in ADSCs in co-transfection groups |
|
|
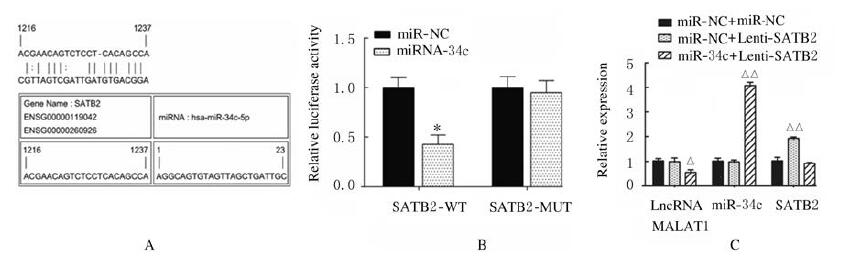
miR-34c与SATB2结合位点见图 11A。荧光素酶报告实验结果显示:与miR-NC组比较,miR-34c转染后SATB2-WT组荧光素酶活性明显降低(P < 0.01),SATB2-MUT组荧光素酶活性差异无统计学意义(P > 0.05),见图 11B;与miR-NC+Lenti-NC组比较,miR-NC+Lenti-SATB2组ADSCs中SATB2 mRNA表达水平明显升高(P < 0.01),见图 11C;Runx2、OPN和OCN蛋白表达水平明显升高(P < 0.01),见图 12;ALP和ARS水平明显升高(P < 0.01),见图 13(插页三)和图 14;miR-34c+Lenti-SATB2组LncRNA MALAT1表达水平明显降低(P < 0.05),miR-34c表达水平明显升高(P < 0.01),见图 11C;SATB2、Runx2、OPN和OCN蛋白表达水平及ALP和ARS水平差异无统计学意义(P > 0.05)。
|
| *P < 0.01 vs miR-NC group; △P < 0.05, △△P < 0.01 vs Lenti-NC + miR-NC group. 图 11 miR-34c与SATB2结合位点和靶向结合(A)及各组细胞中LncRNA MALAT1、miR-34c和SATB2 mRNA表达水平(B, C) Fig. 11 Binding sites of miR-34c and SATB2 and targeted binding (A) and expression levels of LncRNA MALAT1, miR-34c and SATB2 mRNA in cells in various groups(B, C) |
|
|

|
| Lane 1: miR-NC+Lenti-NC group; Lane 2: miR-NC+ Lenti-SATB2 group; Lane 3: miR-34c+ Lenti-SATB2 group.*P < 0.01 vs miR-NC+Lenti-NC group. 图 12 Western blotting法检测各组ADSCs中Runx2、OPN和OCN蛋白表达电泳图(A)和直条图(B) Fig. 12 Electrophoregram (A) and histogram (B) of expressions of Runx2, OPN and OCN proteins in ADSCs in various groups detected by Western blotting method |
|
|

|
| 图 13 ALP和ARS染色检测ADSCs中ALP活性和钙盐沉积 Fig. 13 ALP activities and calcium salt deposition in ADSCs detected by ALP and ARS staining |
|
|

|
| *P < 0.01 vs miR-NC+Lenti-NC group. 图 14 各组ADSCs中ALP和ARS水平 Fig. 14 Levels of ALP and ARS in ADSCs in various groups |
|
|
ADSCs是位于脂肪组织中的再生细胞,具有多向分化潜能[11]。ADSCs可通过脂肪细胞置换促进组织新陈代谢和血管生成[12]。BMSCs是骨再生的细胞来源,最近研究[13]表明:BMSCs的增殖能力和成骨分化能力随年龄和骨质疏松程度的增加而降低,而ADSCs在老年和骨质疏松情况下成骨分化能力不变。因此,ADSCs成骨分化机制受到越来越多的关注。LncRNAs是一种转录本长度超过200nt的非编码RNA,参与多种生物学过程,包括表观遗传调控、RNA衰变、细胞分化和转录[14]。研究[15]表明:LncRNA在调节骨骼系统相关的生物学活性方面(骨质疏松症和骨关节炎)具有重要作用。LncRNA MALAT1通过调节miR-214/ATF4轴促进成骨分化[16]。
研究[17]表明:LncRNA在成骨-成脂谱系中起表观遗传调控作用。LncRNA MIAT1促进ADSCs的成骨分化并逆转炎症的不良反应[18]。本研究结果显示:在ADSCs成骨分化过程中,MALAT1的表达水平升高,MALAT1正向调控ADSCs成骨分化,这与之前的研究结果一致。研究[19]表明:miRNA参与调节成骨细胞和破骨细胞分化过程,如miR-221-5p通过靶向smad3抑制BMSCs成骨分化;miRNA-429通过靶向SCD-1抑制氧化应激下人BMSCs的成骨分化[20]。lncRNA和miRNA间的相互作用参与成骨分化过程,GAO等[21]研究表明:骨质疏松患者的BMSCs中MALAT1表达水平明显降低,成骨诱导BMSCs后MALAT1表达水平升高。敲除MALAT1则抑制BMSCs的成骨分化,MALAT1通过靶向miR-143促进BMSCs的成骨分化;lncRNA TUG1通过使miR-204-5p海绵化而促进成骨细胞的分化,从而导致Runx2表达上调[22]。LncRNA MEG3通过靶向miR-133a-3p抑制BMSCs的成骨分化[23]。本研究结果显示:在ADSCs成骨分化过程中,miR-34c与MALAT1间存在相互调控作用,MALAT1通过靶向抑制miR-34c的表达促进ADSCs成骨分化。
SATB2是成骨细胞分化的特异性免疫组织化学生物标记物,对骨和软组织肿瘤发挥重要作用[24]。研究[25]表明:许多特异性miRNAs通过转化生长因子β(TGF-β)/骨形态发生蛋白(BMP)信号途径参与SATB2诱导的早期成骨分化。miR-34b/c靶向调控SATB2[26-27]。miR-383通过靶向SATB2负调控大鼠BMSCs的成骨细胞分化[28]。本研究结果显示:SATB2可促进ADSCs成骨分化,miR-34c通过靶向SATB2负调控ADSCs成骨分化。
综上所述,LncRNA MALAT1通过microRNA-34c/SATB2轴促进ADSCs的成骨分化。本研究结果为探究lncRNA调控ADSCs成骨分化的机制和骨损伤的治疗提供了新的科学依据。
| [1] |
MADHURAKKAT PERIKAMANA S K, LEE J, AHMAD T, et al. Harnessing biochemical and structural cues for tenogenic differentiation of adipose derived stem cells (ADSCs) and development of an in vitro tissue interface mimicking tendon-bone insertion graft[J]. Biomaterials, 2018, 165: 79-93. DOI:10.1016/j.biomaterials.2018.02.046 |
| [2] |
FERNANDES M, VALENTE S G, SABONGI R G, et al. Bone marrow-derived mesenchymal stem cells versus adipose-derived mesenchymal stem cells for peripheral nerve regeneration[J]. Neural Regen Res, 2018, 13(1): 100-104. DOI:10.4103/1673-5374.224378 |
| [3] |
宋孟晓, 王燕, 刘进忠. miR-222-5p在人根尖乳头干细胞成骨/成牙本质向分化中的作用[J]. 山东大学学报(医学版), 2020, 58(3): 87-93, 112. |
| [4] |
PENG S P, CAO L H, HE S W, et al. An overview of long noncoding RNAs involved in bone regeneration from mesenchymal stem cells[J]. Stem Cells Int, 2018, 2018: 8273648. |
| [5] |
GAO X R, GE J, LI W Y, et al. LncRNA KCNQ1OT1 promotes osteogenic differentiation to relieve osteolysis via Wnt/β-catenin activation[J]. Cell Biosci, 2018, 8: 19. DOI:10.1186/s13578-018-0216-4 |
| [6] |
HUANG G X, KANG Y, HUANG Z Y, et al. Identification and characterization of long non-coding RNAs in osteogenic differentiation of human adipose-derived stem cells[J]. Cell Physiol Biochem, 2017, 42(3): 1037-1050. DOI:10.1159/000478751 |
| [7] |
XIAO X X, ZHOU T W, GUO S C, et al. LncRNA MALAT1 sponges miR-204 to promote osteoblast differentiation of human aortic valve interstitial cells through up-regulating Smad4[J]. Int J Cardiol, 2017, 243: 404-412. DOI:10.1016/j.ijcard.2017.05.037 |
| [8] |
NARAYANAN A, SRINAATH N, ROHINI M, et al. Regulation of Runx2 by MicroRNAs in osteoblast differentiation[J]. Life Sci, 2019, 232: 116676. DOI:10.1016/j.lfs.2019.116676 |
| [9] |
BAE Y J, YANG T, ZENG H C, et al. miRNA-34c regulates Notch signaling during bone development[J]. Hum Mol Genet, 2012, 21(13): 2991-3000. DOI:10.1093/hmg/dds129 |
| [10] |
YU L J, XU Y S, QU H, et al. Decrease of MiR-31 induced by TNF-α inhibitor activates SATB2/RUNX2 pathway and promotes osteogenic differentiation in ethanol-induced osteonecrosis[J]. J Cell Physiol, 2019, 234(4): 4314-4326. DOI:10.1002/jcp.27210 |
| [11] |
王洋, 宋卓悦, 连晓磊, 等. 脂肪和滑膜来源间充质干细胞成软骨分化能力的比较[J]. 郑州大学学报(医学版), 2019, 54(3): 394-398. |
| [12] |
NADERI N, COMBELLACK E J, GRIFFIN M, et al. The regenerative role of adipose-derived stem cells (ADSC) in plastic and reconstructive surgery[J]. Int Wound J, 2017, 14(1): 112-124. DOI:10.1111/iwj.12569 |
| [13] |
AGHEBATI-MALEKI L, DOLATI S, ZANDI R, et al. Prospect of mesenchymal stem cells in therapy of osteoporosis:A review[J]. J Cell Physiol, 2019, 234(6): 8570-8578. DOI:10.1002/jcp.27833 |
| [14] |
RANSOHOFF J D, WEI Y N, KHAVARI P A. The functions and unique features of long intergenic non-coding RNA[J]. Nat Rev Mol Cell Biol, 2018, 19(3): 143-157. DOI:10.1038/nrm.2017.104 |
| [15] |
HUYNH N P, ANDERSON B A, GUILAK F, et al. Emerging roles for long noncoding RNAs in skeletal biology and disease[J]. Connect Tissue Res, 2017, 58(1): 116-141. DOI:10.1080/03008207.2016.1194406 |
| [16] |
HUANG X Z, HUANG J, LI W Z, et al. LncRNA-MALAT1 promotes osteogenic differentiation through regulating ATF4 by sponging miR-214:Implication of steroid-induced avascular necrosis of the femoral head[J]. Steroids, 2020, 154: 108533. DOI:10.1016/j.steroids.2019.108533 |
| [17] |
YOSHIOKA H, YOSHIKO Y. The roles of long non-protein-coding RNAs in osteo-adipogenic lineage commitment[J]. Int J Mol Sci, 2017, 18(6): E1236. DOI:10.3390/ijms18061236 |
| [18] |
JIN C Y, ZHENG Y F, HUANG Y P, et al. Long non-coding RNA MIAT knockdown promotes osteogenic differentiation of human adipose-derived stem cells[J]. Cell Biol Int, 2017, 41(1): 33-41. |
| [19] |
FAN F Y, DENG R, LAI S H, et al. Inhibition of microRNA-221-5p induces osteogenic differentiation by directly targeting smad3 in myeloma bone disease mesenchymal stem cells[J]. Oncol Lett, 2019, 18(6): 6536-6544. |
| [20] |
LAN C G, LONG L Z, XIE K G, et al. MiRNA-429 suppresses osteogenic differentiation of human adipose-derived mesenchymal stem cells under oxidative stress via targeting SCD-1[J]. Exp Ther Med, 2020, 19(1): 696-702. |
| [21] |
GAO Y, XIAO F, WANG C L, et al. Long noncoding RNA MALAT1 promotes osterix expression to regulate osteogenic differentiation by targeting miRNA-143 in human bone marrow-derived mesenchymal stem cells[J]. J Cell Biochem, 2018, 119(8): 6986-6996. DOI:10.1002/jcb.26907 |
| [22] |
YU C, LI L F, XIE F, et al. LncRNA TUG1 sponges miR-204-5p to promote osteoblast differentiation through upregulating Runx2 in aortic valve calcification[J]. Cardiovasc Res, 2018, 114(1): 168-179. DOI:10.1093/cvr/cvx180 |
| [23] |
WANG Q J, LI Y X, ZHANG Y, et al. LncRNA MEG3 inhibited osteogenic differentiation of bone marrow mesenchymal stem cells from postmenopausal osteoporosis by targeting miR-133a-3p[J]. Biomed Pharmacother, 2017, 89: 1178-1186. DOI:10.1016/j.biopha.2017.02.090 |
| [24] |
ZARATE Y A, STEINRATHS M, MATTHEWS A, et al. Bone health and SATB2-associated syndrome[J]. Clin Genet, 2018, 93(3): 588-594. DOI:10.1111/cge.13121 |
| [25] |
GONG Y M, XU F, ZHANG L, et al. MicroRNA expression signature for Satb2-induced osteogenic differentiation in bone marrow stromal cells[J]. Mol Cell Biochem, 2014, 387(1/2): 227-239. |
| [26] |
王亮, 钟武, 陈睦虎. miR-34a对人脂肪间充质干细胞成骨向分化的调控[J]. 西安交通大学学报(医学版), 2018, 39(5): 675-679, 708. |
| [27] |
HAO J B, ZHANG L, CONG G T, et al. MicroRNA-34b/c inhibits aldosterone-induced vascular smooth muscle cell calcification via a SATB2/Runx2 pathway[J]. Cell Tissue Res, 2016, 366(3): 733-746. DOI:10.1007/s00441-016-2469-8 |
| [28] |
TANG J F, ZHANG Z, JIN X Y, et al. miR-383 negatively regulates osteoblastic differentiation of bone marrow mesenchymal stem cells in rats by targeting Satb2[J]. Bone, 2018, 114: 137-143. DOI:10.1016/j.bone.2018.06.010 |
 2020, Vol. 46
2020, Vol. 46
